Introduction
Realistic prediction of protein surface regions that are preferentially recognized by antibodies (antigenic epitopes) can help the design of vaccine components and immuno-diagnostic reagents.
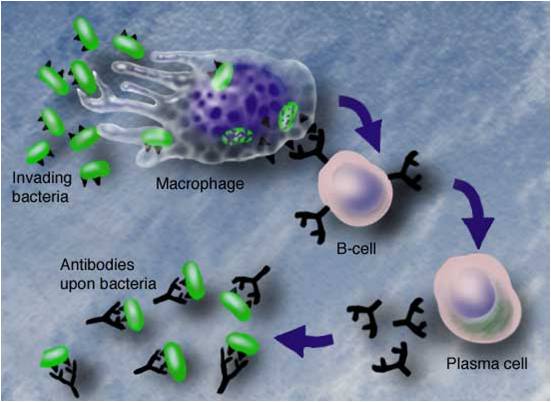 By secreting antibodies against antigens, B-cells play an important role in immune system to fight against the invasive pathogenic organism or substances. Antigenic epitopes are regions of protein surface that are preferentially recognized by B-cell antibodies . Prediction of antigenic epitopes is useful for the investigation to the mechanism in body self-protection systems and help during the design of vaccine components and immuno-diagnostic reagents.
By secreting antibodies against antigens, B-cells play an important role in immune system to fight against the invasive pathogenic organism or substances. Antigenic epitopes are regions of protein surface that are preferentially recognized by B-cell antibodies . Prediction of antigenic epitopes is useful for the investigation to the mechanism in body self-protection systems and help during the design of vaccine components and immuno-diagnostic reagents.
Usually, B-cell antigenic epitopes are classified as either continuous or discontinuous. A continuous (also called linear) epitope is a consecutive fragment from the protein sequence, and a discontinuous epitope is composed of several fragments scattered along the protein sequence, which form the antigen-binding interface. The boundary between continuous and discontinuous epitopes is vague; a fragment in a discontinuous epitope can be considered as a continuous epitope.
Currently, the majority of available epitope prediction methods focus on continuous epitopes due to the convenience of the investigation in which the amino acid sequence of a protein is taken as the input. Such prediction methods are based upon the amino acid properties including hydrophilicity , solvent accessibility , secondary structure , flexibility , and antigenicity . In addition, based on the known linear epitope databases such as Bcipep and FIMM , there also exist some methods using machine learning algorithms such as Hidden Markov Model (HMM) , Artificial Neural Network (ANN) , and Support Vector Machine (SVM) to locate linear epitopes.
In this work, we developed a new method to predict antigenic epitope with lastest sequence input from IEDB database. In our method, Support Vector Machine (SVM) has been utilized by combining the Tri-peptide similarity and Propensity scores (SVMTriP) in order to achieve the better prediction performance.
Citation: B. Yao, L. Zhang, S. Liang, C. Zhang. SVMTriP: A method to predict antigenic epitopes using support vector machine to integrate tri-peptide similarity and propensity. PLoS ONE (2012); 7(9):e45152.
