
| Microexon ID | Zm_9:155241201-155241215:- |
| Species | Zea mays | Coordinates | 9:155241201..155241215 |
| Microexon Cluster ID | MEP45 |
| Size | 15 |
| Phase | 2 |
| Pfam Domain Motif | RPE65 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 47,15,46 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | AGRGTTGGKCCTAAYCCMAAGTTTGYYCCWGTKGCTGGATAYCAYTGGTTTGATGGAGATGGMATGATTCATGSYWTGCGYATYAAAGATGGAAAAGCWACWTATGTY |
| Logo of Microexon-tag DNA Seq | 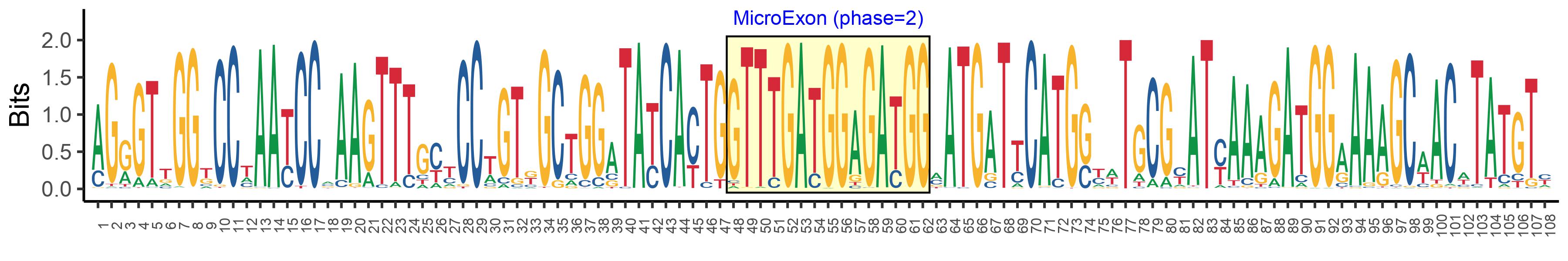 |
| Alignment of exons | 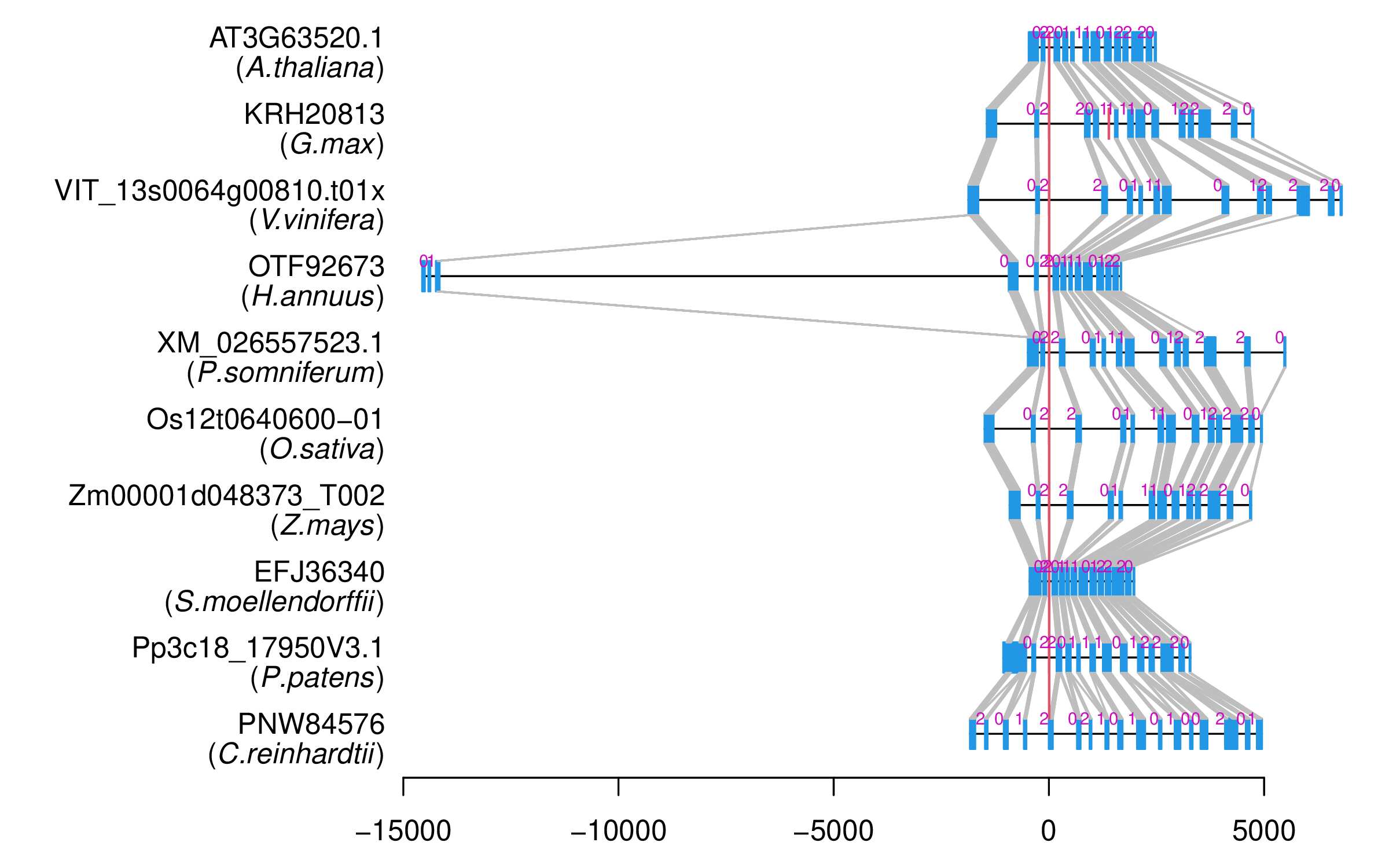 |
| Microexon DNA seq | GTTTGATGGAGACGG |
| Microexon Amino Acid seq | WFDGDG |
| Microexon-tag DNA Seq | AGGGTTGGGCCTAATCCGAAGTTTGCTCCTGTTGCGGGGTATCACTGGTTTGATGGAGACGGGATGATTCATGCCATGCGTATTAAGGATGGAAAAGCTACCTATGTA |
| Microexon-tag Amino Acid Seq | RVGPNPKFAPVAGYHWFDGDGMIHAMRIKDGKATYV |
| Microexon-tag spanning region | 155240729-155241475 |
| Microexon-tag prediction score | 0.9765 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | Zm00001d048373_T002x |
| Reference Transcript ID | Zm00001d048373_T002 |
| Gene ID | Zm00001d048373 |
| Gene Name | white cap1 |
| Transcript ID | Zm00001d048373_T002 |
| Protein ID | Zm00001d048373_P002 |
| Gene ID | Zm00001d048373 |
| Gene Name | white cap1 |
| Pfam domain motif | RPE65 |
| Motif E-value | 2.9e-128 |
| Motif start | 60 |
| Motif end | 538 |
| Protein seq | >Zm00001d048373_P002 MGTEAEQPDMDSHRNDGVVVVPAPRPRKGLASWALDLLESLAVRLGHDKTKPLHWLSGNFAPVVEETPPAPNLSVRGHLP ECLNGEFVRVGPNPKFAPVAGYHWFDGDGMIHAMRIKDGKATYVSRYVKTARLKQEEYFGGAKFMKIGDLKGFFGLFMVQ MQQLRKKFKVLDFTYGFGTANTALIYHHGKLMALSEADKPYVVKVLEDGDLQTLGLLDYDKRLKHSFTAHPKVDPFTDEM FTFGYSHEPPYCTYRVINKEGAMLDPVPITIPESVMMHDFAITENYSIFMDLPLLFRPKEMVKNGEFIYKFDPTKKGRFG ILPRYAKDDKLIRWFQLPNCFIFHNANAWEEGDEVVLITCRLENPDLDKVNGYQSDKLENFGNELYEMRFNMKTGAASQK QLSVSAVDFPRVNESYTGRKQRYVYCTILDSIAKVTGIIKFDLHAEPESGVKELEVGGNVQGIYDLGPGRFGSEAIFVPK HPGVSGEEDDGYLIFFVHDENTGKSEVNVIDAKTMSADPVAVVELPNRVPYGFHAFFVTEDQLARQAEGQ* |
| CDS seq | >Zm00001d048373_T002 ATGGGGACGGAGGCGGAGCAGCCGGACATGGACAGCCACCGAAACGACGGCGTCGTGGTGGTGCCAGCGCCGCGCCCGCG TAAGGGGCTCGCCTCCTGGGCGCTTGACCTGCTCGAGTCCCTCGCCGTGCGCCTCGGCCACGACAAGACCAAGCCGCTCC ACTGGCTCTCCGGCAACTTCGCCCCCGTCGTCGAGGAGACCCCGCCGGCCCCAAACCTTAGCGTCCGCGGACACCTCCCG GAGTGCTTGAATGGAGAGTTTGTCAGGGTTGGGCCTAATCCGAAGTTTGCTCCTGTTGCGGGGTATCACTGGTTTGATGG AGACGGGATGATTCATGCCATGCGTATTAAGGATGGAAAAGCTACCTATGTATCAAGATATGTGAAGACTGCCCGCCTCA AACAAGAGGAGTATTTTGGTGGAGCAAAGTTTATGAAGATTGGAGACCTTAAGGGATTTTTTGGATTGTTTATGGTCCAA ATGCAGCAACTTCGGAAAAAATTCAAAGTCTTGGATTTTACCTATGGATTTGGGACAGCTAATACTGCACTTATATATCA TCATGGTAAACTCATGGCCTTGTCAGAAGCAGATAAGCCATATGTTGTTAAGGTCCTTGAAGATGGAGACTTGCAGACTC TTGGCTTGTTGGATTATGACAAAAGGTTGAAACATTCTTTTACTGCCCATCCAAAGGTTGACCCTTTTACAGATGAAATG TTCACATTCGGATATTCACATGAACCTCCATACTGTACATACCGTGTGATTAACAAAGAAGGAGCTATGCTTGATCCTGT GCCAATAACAATACCGGAATCTGTAATGATGCATGATTTTGCCATCACAGAGAATTACTCTATTTTTATGGACCTCCCTT TATTGTTCCGACCAAAGGAAATGGTGAAGAACGGTGAGTTTATCTACAAGTTTGATCCTACAAAGAAAGGTCGTTTTGGT ATTCTCCCCCGCTATGCAAAGGATGACAAACTCATCAGATGGTTTCAACTCCCTAATTGTTTCATATTCCATAATGCTAA TGCTTGGGAAGAGGGTGATGAAGTTGTTCTCATTACCTGCCGCCTTGAGAATCCAGATTTGGACAAGGTGAATGGATATC AAAGTGACAAGCTCGAAAACTTCGGGAATGAGCTGTACGAGATGAGATTCAACATGAAAACGGGTGCTGCTTCACAAAAG CAATTGTCTGTTTCTGCTGTGGATTTTCCTCGTGTTAATGAGAGCTATACTGGCAGAAAGCAGCGGTATGTCTACTGCAC TATACTTGACAGCATTGCGAAGGTGACTGGCATCATAAAGTTTGATCTGCATGCTGAACCGGAAAGTGGTGTGAAAGAAC TTGAAGTGGGAGGAAATGTACAAGGCATATATGACCTGGGACCTGGTAGATTTGGTTCAGAGGCGATTTTTGTTCCCAAG CATCCAGGTGTGTCCGGAGAAGAAGATGACGGCTATTTGATATTCTTTGTACACGACGAGAATACAGGGAAATCTGAAGT AAATGTTATCGATGCAAAGACAATGTCTGCTGATCCAGTTGCGGTGGTTGAGCTTCCTAATAGGGTTCCTTATGGATTCC ATGCCTTCTTTGTAACTGAGGACCAACTGGCTCGACAGGCGGAGGGGCAGTGA |
| Microexon DNA seq | GTTTGATGGAGACGG |
| Microexon Amino Acid seq | WFDGDG |
| Microexon-tag DNA Seq | AGGGTTGGGCCTAATCCGAAGTTTGCTCCTGTTGCGGGGTATCACTGGTTTGATGGAGACGGGATGATTCATGCCATGCGTATTAAGGATGGAAAAGCTACCTATGTA |
| Microexon-tag Amino Acid seq | RVGPNPKFAPVAGYHWFDGDGMIHAMRIKDGKATYV |
| Transcript ID | Zm00001d048373_T002 |
| Gene ID | Zm.38699 |
| Gene Name | white cap1 |
| Pfam domain motif | RPE65 |
| Motif E-value | 3e-128 |
| Motif start | 60 |
| Motif end | 538 |
| Protein seq | >Zm00001d048373_T002 MGTEAEQPDMDSHRNDGVVVVPAPRPRKGLASWALDLLESLAVRLGHDKTKPLHWLSGNFAPVVEETPPAPNLSVRGHLP ECLNGEFVRVGPNPKFAPVAGYHWFDGDGMIHAMRIKDGKATYVSRYVKTARLKQEEYFGGAKFMKIGDLKGFFGLFMVQ MQQLRKKFKVLDFTYGFGTANTALIYHHGKLMALSEADKPYVVKVLEDGDLQTLGLLDYDKRLKHSFTAHPKVDPFTDEM FTFGYSHEPPYCTYRVINKEGAMLDPVPITIPESVMMHDFAITENYSIFMDLPLLFRPKEMVKNGEFIYKFDPTKKGRFG ILPRYAKDDKLIRWFQLPNCFIFHNANAWEEGDEVVLITCRLENPDLDKVNGYQSDKLENFGNELYEMRFNMKTGAASQK QLSVSAVDFPRVNESYTGRKQRYVYCTILDSIAKVTGIIKFDLHAEPESGVKELEVGGNVQGIYDLGPGRFGSEAIFVPK HPGVSGEEDDGYLIFFVHDENTGKSEVNVIDAKTMSADPVAVVELPNRVPYGFHAFFVTEDQLARQAEGQ* |
| CDS seq | >Zm00001d048373_T002 ATGGGGACGGAGGCGGAGCAGCCGGACATGGACAGCCACCGAAACGACGGCGTCGTGGTGGTGCCAGCGCCGCGCCCGCG TAAGGGGCTCGCCTCCTGGGCGCTTGACCTGCTCGAGTCCCTCGCCGTGCGCCTCGGCCACGACAAGACCAAGCCGCTCC ACTGGCTCTCCGGCAACTTCGCCCCCGTCGTCGAGGAGACCCCGCCGGCCCCAAACCTTAGCGTCCGCGGACACCTCCCG GAGTGCTTGAATGGAGAGTTTGTCAGGGTTGGGCCTAATCCGAAGTTTGCTCCTGTTGCGGGGTATCACTGGTTTGATGG AGACGGGATGATTCATGCCATGCGTATTAAGGATGGAAAAGCTACCTATGTATCAAGATATGTGAAGACTGCCCGCCTCA AACAAGAGGAGTATTTTGGTGGAGCAAAGTTTATGAAGATTGGAGACCTTAAGGGATTTTTTGGATTGTTTATGGTCCAA ATGCAGCAACTTCGGAAAAAATTCAAAGTCTTGGATTTTACCTATGGATTTGGGACAGCTAATACTGCACTTATATATCA TCATGGTAAACTCATGGCCTTGTCAGAAGCAGATAAGCCATATGTTGTTAAGGTCCTTGAAGATGGAGACTTGCAGACTC TTGGCTTGTTGGATTATGACAAAAGGTTGAAACATTCTTTTACTGCCCATCCAAAGGTTGACCCTTTTACAGATGAAATG TTCACATTCGGATATTCACATGAACCTCCATACTGTACATACCGTGTGATTAACAAAGAAGGAGCTATGCTTGATCCTGT GCCAATAACAATACCGGAATCTGTAATGATGCATGATTTTGCCATCACAGAGAATTACTCTATTTTTATGGACCTCCCTT TATTGTTCCGACCAAAGGAAATGGTGAAGAACGGTGAGTTTATCTACAAGTTTGATCCTACAAAGAAAGGTCGTTTTGGT ATTCTCCCCCGCTATGCAAAGGATGACAAACTCATCAGATGGTTTCAACTCCCTAATTGTTTCATATTCCATAATGCTAA TGCTTGGGAAGAGGGTGATGAAGTTGTTCTCATTACCTGCCGCCTTGAGAATCCAGATTTGGACAAGGTGAATGGATATC AAAGTGACAAGCTCGAAAACTTCGGGAATGAGCTGTACGAGATGAGATTCAACATGAAAACGGGTGCTGCTTCACAAAAG CAATTGTCTGTTTCTGCTGTGGATTTTCCTCGTGTTAATGAGAGCTATACTGGCAGAAAGCAGCGGTATGTCTACTGCAC TATACTTGACAGCATTGCGAAGGTGACTGGCATCATAAAGTTTGATCTGCATGCTGAACCGGAAAGTGGTGTGAAAGAAC TTGAAGTGGGAGGAAATGTACAAGGCATATATGACCTGGGACCTGGTAGATTTGGTTCAGAGGCGATTTTTGTTCCCAAG CATCCAGGTGTGTCCGGAGAAGAAGATGACGGCTATTTGATATTCTTTGTACACGACGAGAATACAGGGAAATCTGAAGT AAATGTTATCGATGCAAAGACAATGTCTGCTGATCCAGTTGCGGTGGTTGAGCTTCCTAATAGGGTTCCTTATGGATTCC ATGCCTTCTTTGTAACTGAGGACCAACTGGCTCGACAGGCGGAGGGGCAGTGA |