
| Microexon ID | At_3:23453406-23453420:+ |
| Species | Arabidopsis thaliana | Coordinates | 3:23453406..23453420 |
| Microexon Cluster ID | MEP45 |
| Size | 15 |
| Phase | 2 |
| Pfam Domain Motif | RPE65 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 47,15,46 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | AGRGTTGGKCCTAAYCCMAAGTTTGYYCCWGTKGCTGGATAYCAYTGGTTTGATGGAGATGGMATGATTCATGSYWTGCGYATYAAAGATGGAAAAGCWACWTATGTY |
| Logo of Microexon-tag DNA Seq | 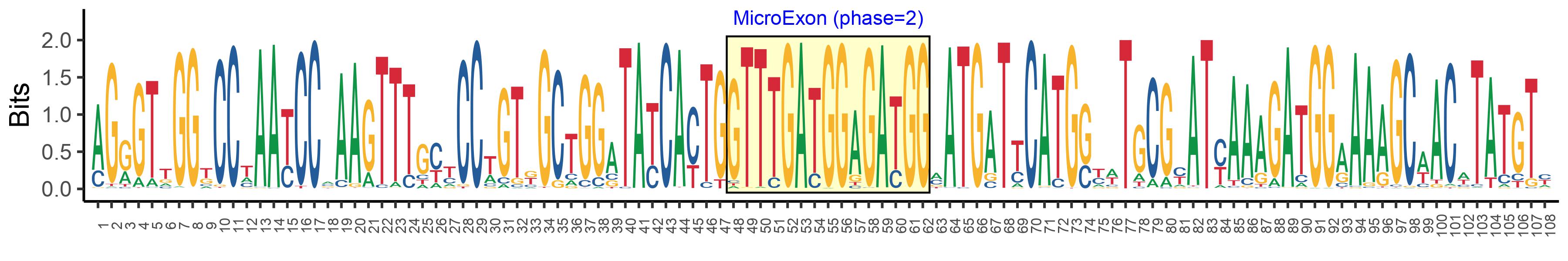 |
| Alignment of exons | 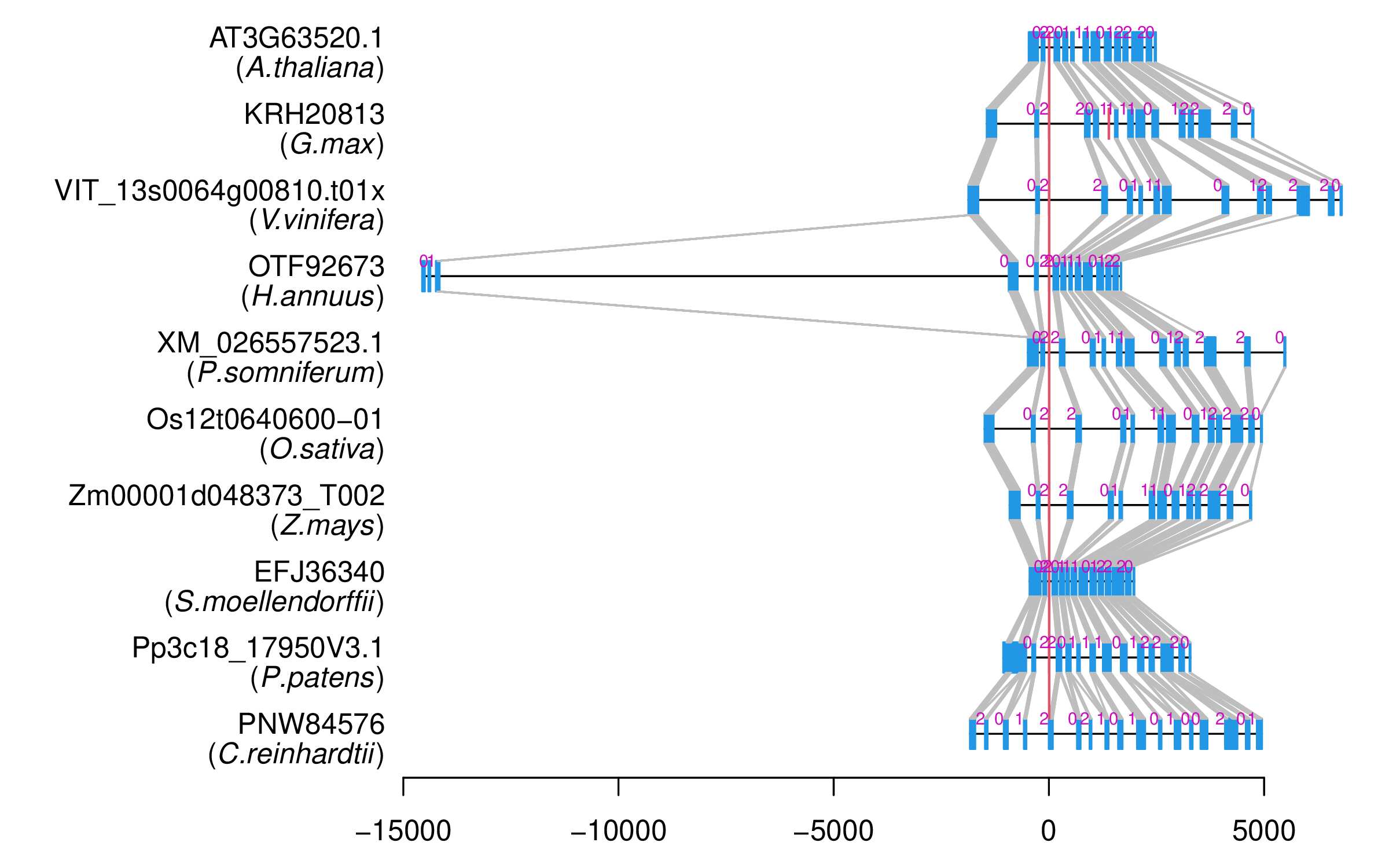 |
| Microexon DNA seq | GTTTGATGGAGATGG |
| Microexon Amino Acid seq | WFDGDG |
| Microexon-tag DNA Seq | AGGGTTGGTCCAAACCCCAAGTTTGATGCTGTCGCTGGATATCACTGGTTTGATGGAGATGGGATGATTCATGGGGTACGCATCAAAGATGGGAAAGCTACTTATGTT |
| Microexon-tag Amino Acid Seq | RVGPNPKFDAVAGYHWFDGDGMIHGVRIKDGKATYV |
| Microexon-tag spanning region | 23453261-23453582 |
| Microexon-tag prediction score | 0.9719 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | AT3G63520.1x |
| Reference Transcript ID | AT3G63520.1 |
| Gene ID | AT3G63520 |
| Gene Name | CCD1 |
| Transcript ID | AT3G63520.1 |
| Protein ID | AT3G63520.1 |
| Gene ID | AT3G63520 |
| Gene Name | CCD1 |
| Pfam domain motif | RPE65 |
| Motif E-value | 1.7e-133 |
| Motif start | 52 |
| Motif end | 527 |
| Protein seq | >AT3G63520.1 MAEKLSDGSSIISVHPRPSKGFSSKLLDLLERLVVKLMHDASLPLHYLSGNFAPIRDETPPVKDLPVHGFLPECLNGEFV RVGPNPKFDAVAGYHWFDGDGMIHGVRIKDGKATYVSRYVKTSRLKQEEFFGAAKFMKIGDLKGFFGLLMVNVQQLRTKL KILDNTYGNGTANTALVYHHGKLLALQEADKPYVIKVLEDGDLQTLGIIDYDKRLTHSFTAHPKVDPVTGEMFTFGYSHT PPYLTYRVISKDGIMHDPVPITISEPIMMHDFAITETYAIFMDLPMHFRPKEMVKEKKMIYSFDPTKKARFGVLPRYAKD ELMIRWFELPNCFIFHNANAWEEEDEVVLITCRLENPDLDMVSGKVKEKLENFGNELYEMRFNMKTGSASQKKLSASAVD FPRINECYTGKKQRYVYGTILDSIAKVTGIIKFDLHAEAETGKRMLEVGGNIKGIYDLGEGRYGSEAIYVPRETAEEDDG YLIFFVHDENTGKSCVTVIDAKTMSAEPVAVVELPHRVPYGFHALFVTEEQLQEQTLI* |
| CDS seq | >AT3G63520.1 ATGGCGGAGAAACTCAGTGATGGCAGCAGCATCATCTCAGTCCATCCTAGACCCTCCAAGGGTTTCTCCTCGAAGCTTCT CGATCTTCTCGAGAGACTTGTTGTCAAGCTCATGCACGATGCTTCTCTCCCTCTCCACTACCTCTCAGGCAACTTCGCTC CCATCCGTGATGAAACTCCTCCCGTCAAGGATCTCCCCGTCCATGGATTTCTTCCCGAATGCTTGAATGGTGAATTTGTG AGGGTTGGTCCAAACCCCAAGTTTGATGCTGTCGCTGGATATCACTGGTTTGATGGAGATGGGATGATTCATGGGGTACG CATCAAAGATGGGAAAGCTACTTATGTTTCTCGATATGTTAAGACATCACGTCTTAAGCAGGAAGAGTTCTTCGGAGCTG CCAAATTCATGAAGATTGGTGACCTTAAGGGGTTTTTCGGATTGCTAATGGTCAATGTCCAACAGCTGAGAACGAAGCTC AAAATATTGGACAACACTTATGGAAATGGAACTGCCAATACAGCACTCGTATATCACCATGGAAAACTTCTAGCATTACA GGAGGCAGATAAGCCGTACGTCATCAAAGTTTTGGAAGATGGAGACCTGCAAACTCTTGGTATAATAGATTATGACAAGA GATTGACCCACTCCTTCACTGCTCACCCAAAAGTTGACCCGGTTACGGGTGAAATGTTTACATTCGGCTATTCGCATACG CCACCTTATCTCACATACAGAGTTATCTCGAAAGATGGCATTATGCATGACCCAGTCCCAATTACTATATCAGAGCCTAT CATGATGCATGATTTTGCTATTACTGAGACTTATGCAATCTTCATGGATCTTCCTATGCACTTCAGGCCAAAGGAAATGG TGAAAGAGAAGAAAATGATATACTCATTTGATCCCACAAAAAAGGCTCGTTTTGGTGTTCTTCCACGCTATGCCAAGGAT GAACTTATGATTAGATGGTTTGAGCTTCCCAACTGCTTTATTTTCCACAACGCCAATGCTTGGGAAGAAGAGGATGAAGT CGTCCTCATCACTTGTCGTCTTGAGAATCCAGATCTTGACATGGTCAGTGGGAAAGTGAAAGAAAAACTCGAAAATTTTG GCAACGAACTGTACGAAATGAGATTCAACATGAAAACGGGCTCAGCTTCTCAAAAAAAACTATCCGCATCTGCGGTTGAT TTCCCCAGAATCAATGAGTGCTACACCGGAAAGAAACAGAGATACGTATATGGAACAATTCTGGACAGTATCGCAAAGGT TACCGGAATCATCAAGTTTGATCTGCATGCAGAAGCTGAGACAGGGAAAAGAATGCTGGAAGTAGGAGGTAATATCAAAG GAATATATGACCTGGGAGAAGGCAGATATGGTTCAGAGGCTATCTATGTTCCGCGTGAGACAGCAGAAGAAGACGACGGT TACTTGATATTCTTTGTTCATGATGAAAACACAGGGAAATCATGCGTGACTGTGATAGACGCAAAAACAATGTCGGCTGA ACCGGTGGCAGTGGTGGAGCTGCCGCACAGGGTCCCATATGGCTTCCATGCCTTGTTTGTTACAGAGGAACAACTCCAGG AACAAACTCTTATATAA |
| Microexon DNA seq | GTTTGATGGAGATGG |
| Microexon Amino Acid seq | WFDGDG |
| Microexon-tag DNA Seq | AGGGTTGGTCCAAACCCCAAGTTTGATGCTGTCGCTGGATATCACTGGTTTGATGGAGATGGGATGATTCATGGGGTACGCATCAAAGATGGGAAAGCTACTTATGTT |
| Microexon-tag Amino Acid seq | RVGPNPKFDAVAGYHWFDGDGMIHGVRIKDGKATYV |
| Transcript ID | At.17447.1 |
| Gene ID | At.17447 |
| Gene Name | CCD1 |
| Pfam domain motif | RPE65 |
| Motif E-value | 1.7e-133 |
| Motif start | 52 |
| Motif end | 527 |
| Protein seq | >At.17447.1 MAEKLSDGSSIISVHPRPSKGFSSKLLDLLERLVVKLMHDASLPLHYLSGNFAPIRDETPPVKDLPVHGFLPECLNGEFV RVGPNPKFDAVAGYHWFDGDGMIHGVRIKDGKATYVSRYVKTSRLKQEEFFGAAKFMKIGDLKGFFGLLMVNVQQLRTKL KILDNTYGNGTANTALVYHHGKLLALQEADKPYVIKVLEDGDLQTLGIIDYDKRLTHSFTAHPKVDPVTGEMFTFGYSHT PPYLTYRVISKDGIMHDPVPITISEPIMMHDFAITETYAIFMDLPMHFRPKEMVKEKKMIYSFDPTKKARFGVLPRYAKD ELMIRWFELPNCFIFHNANAWEEEDEVVLITCRLENPDLDMVSGKVKEKLENFGNELYEMRFNMKTGSASQKKLSASAVD FPRINECYTGKKQRYVYGTILDSIAKVTGIIKFDLHAEAETGKRMLEVGGNIKGIYDLGEGRYGSEAIYVPRETAEEDDG YLIFFVHDENTGKSCVTVIDAKTMSAEPVAVVELPHRVPYGFHALFVTEEQLQEQTLI* |
| CDS seq | >At.17447.1 ATGGCGGAGAAACTCAGTGATGGCAGCAGCATCATCTCAGTCCATCCTAGACCCTCCAAGGGTTTCTCCTCGAAGCTTCT CGATCTTCTCGAGAGACTTGTTGTCAAGCTCATGCACGATGCTTCTCTCCCTCTCCACTACCTCTCAGGCAACTTCGCTC CCATCCGTGATGAAACTCCTCCCGTCAAGGATCTCCCCGTCCATGGATTTCTTCCCGAATGCTTGAATGGTGAATTTGTG AGGGTTGGTCCAAACCCCAAGTTTGATGCTGTCGCTGGATATCACTGGTTTGATGGAGATGGGATGATTCATGGGGTACG CATCAAAGATGGGAAAGCTACTTATGTTTCTCGATATGTTAAGACATCACGTCTTAAGCAGGAAGAGTTCTTCGGAGCTG CCAAATTCATGAAGATTGGTGACCTTAAGGGGTTTTTCGGATTGCTAATGGTCAATGTCCAACAGCTGAGAACGAAGCTC AAAATATTGGACAACACTTATGGAAATGGAACTGCCAATACAGCACTCGTATATCACCATGGAAAACTTCTAGCATTACA GGAGGCAGATAAGCCGTACGTCATCAAAGTTTTGGAAGATGGAGACCTGCAAACTCTTGGTATAATAGATTATGACAAGA GATTGACCCACTCCTTCACTGCTCACCCAAAAGTTGACCCGGTTACGGGTGAAATGTTTACATTCGGCTATTCGCATACG CCACCTTATCTCACATACAGAGTTATCTCGAAAGATGGCATTATGCATGACCCAGTCCCAATTACTATATCAGAGCCTAT CATGATGCATGATTTTGCTATTACTGAGACTTATGCAATCTTCATGGATCTTCCTATGCACTTCAGGCCAAAGGAAATGG TGAAAGAGAAGAAAATGATATACTCATTTGATCCCACAAAAAAGGCTCGTTTTGGTGTTCTTCCACGCTATGCCAAGGAT GAACTTATGATTAGATGGTTTGAGCTTCCCAACTGCTTTATTTTCCACAACGCCAATGCTTGGGAAGAAGAGGATGAAGT CGTCCTCATCACTTGTCGTCTTGAGAATCCAGATCTTGACATGGTCAGTGGGAAAGTGAAAGAAAAACTCGAAAATTTTG GCAACGAACTGTACGAAATGAGATTCAACATGAAAACGGGCTCAGCTTCTCAAAAAAAACTATCCGCATCTGCGGTTGAT TTCCCCAGAATCAATGAGTGCTACACCGGAAAGAAACAGAGATACGTATATGGAACAATTCTGGACAGTATCGCAAAGGT TACCGGAATCATCAAGTTTGATCTGCATGCAGAAGCTGAGACAGGGAAAAGAATGCTGGAAGTAGGAGGTAATATCAAAG GAATATATGACCTGGGAGAAGGCAGATATGGTTCAGAGGCTATCTATGTTCCGCGTGAGACAGCAGAAGAAGACGACGGT TACTTGATATTCTTTGTTCATGATGAAAACACAGGGAAATCATGCGTGACTGTGATAGACGCAAAAACAATGTCGGCTGA ACCGGTGGCAGTGGTGGAGCTGCCGCACAGGGTCCCATATGGCTTCCATGCCTTGTTTGTTACAGAGGAACAACTCCAGG AACAAACTCTTATATAA |