
| Microexon ID | Sm_GL377579:907715-907729:+ |
| Species | Selaginella moellendorffii | Coordinates | GL377579:907715..907729 |
| Microexon Cluster ID | MEP44 |
| Size | 15 |
| Phase | 1 |
| Pfam Domain Motif | Unknown |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 46,15,47 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | RTTTATGRGCTTTGTGATRTTGACTTGAAAGATTTYAGYMTBCAAGCWTWTGGRCARCAAGGBTGTYTACTTCGRAGCTTRCCTRCAGATRTKGTRTTTGACAAYWCW |
| Logo of Microexon-tag DNA Seq | 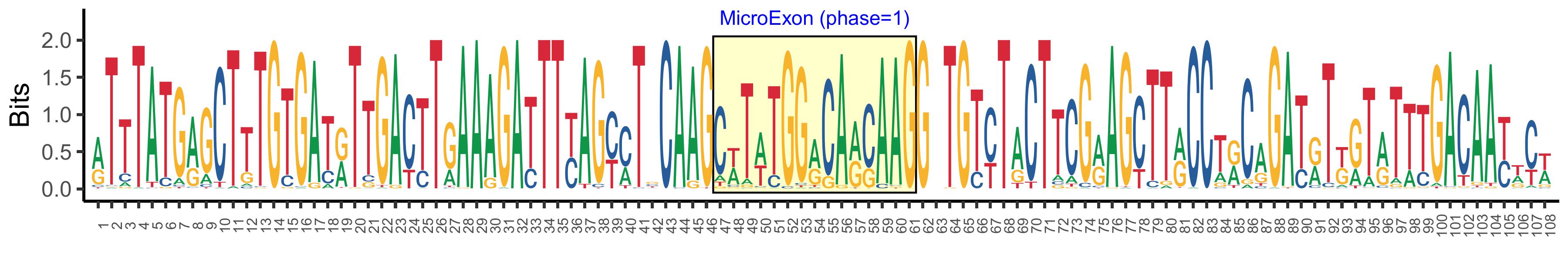 |
| Alignment of exons | 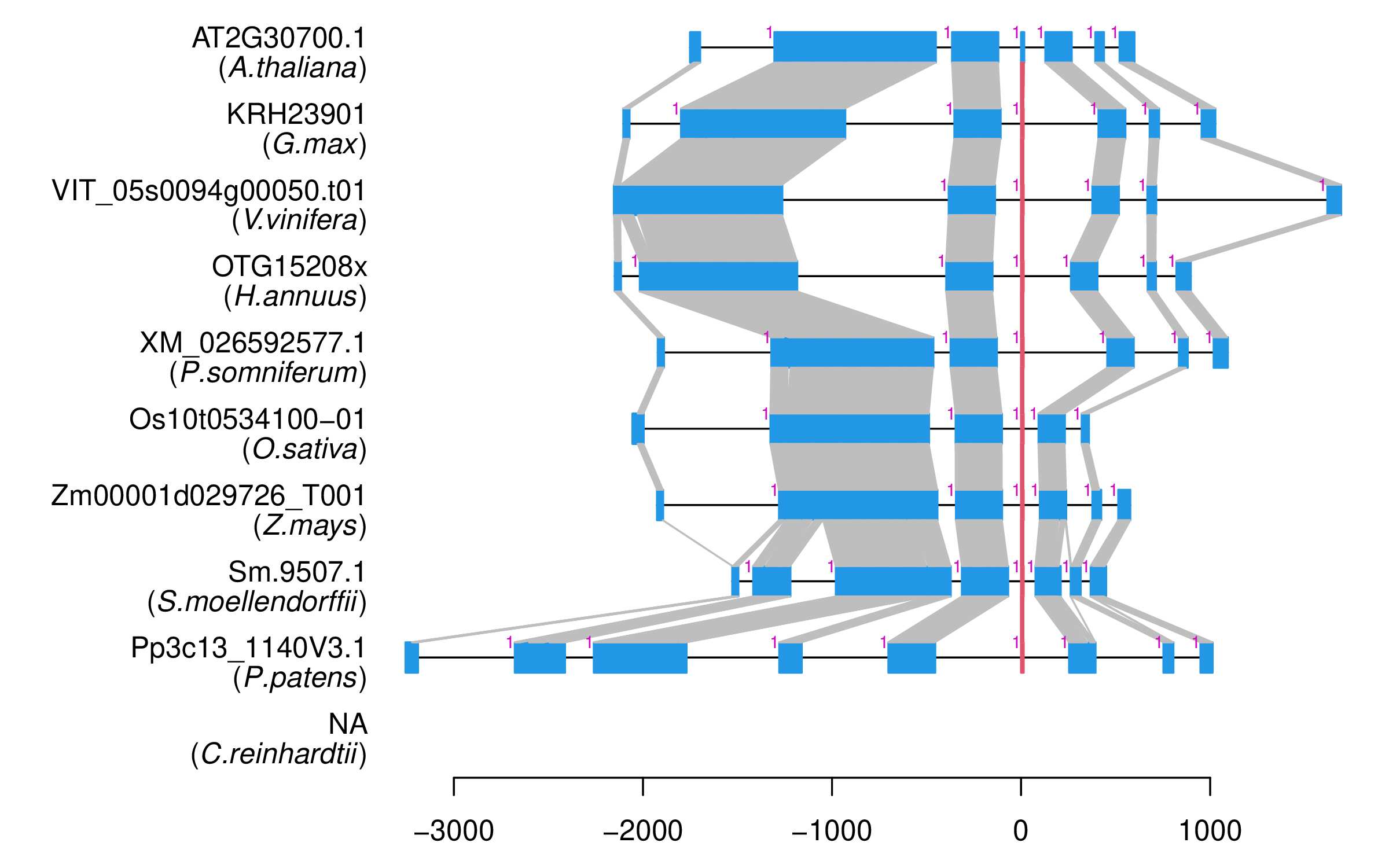 |
Sm_GL377579:907715-907729:+ does not have available information here.
Sm_GL377579:907715-907729:+ does not have available information here.
| Microexon DNA seq | GTGCTGGAGATCAAG |
| Microexon Amino Acid seq | GAGDQG |
| Microexon-tag DNA Seq | ATTTACGATCTATGCGGCATCTATCTCAAAGATTTCAGCCTTCAAGGTGCTGGAGATCAAGGGTGCTTGCTAAATAGCTTGCCGTCTGATGTGATGATCACGAGTAGC |
| Microexon-tag Amino Acid seq | IYDLCGIYLKDFSLQGAGDQGCLLNSLPSDVMITSS |
| Transcript ID | Sm.9507.1 |
| Gene ID | Sm.9507 |
| Gene Name | NA |
| Pfam domain motif | Unknown |
| Motif E-value | NA |
| Motif start | NA |
| Motif end | NA |
| Protein seq | >Sm.9507.1 MAARILAIVLLWLVGFGDLKAEEALPAFDGNLSIPAVVGAQPPFFLPVSPSAPGAIPIVSPSPMIPVLPNPSSPKLSGHC KLNFSALTPAIDRTAQDCLAPLALYVGEVICCPQLQTLFRLAQGHHINTSGRLTFNRTEASYCFSDISSLLVSKGANTTV SEICSMEPKSLTGGQCPVYETKELYRLVNTSRLLPACKGVDPLKECCKPVCQPALEEAALQLASNGSFGFVKQSGLVPAS ADDQVLEDCKDVVLAWVAGQLNLTESNTALRNLFSCKVNKACPLVFSDVSAVVDDCHGLSPSNVTCCSSLHRYISEMQQQ KLITNLQALECVSLLGSMLQKRGVTSNIYDLCGIYLKDFSLQGAGDQGCLLNSLPSDVMITSSSGITFTCDLNDNIAAPW MPMSSSSLCSGGRPNISFPKIPQSAAPLTSGTNFARASSITAFVAPLLILVILSTSI* |
| CDS seq | >Sm.9507.1 ATGGCGGCACGGATCCTTGCCATCGTCTTGCTCTGGCTTGTAGGTTTTGGAGATCTCAAGGCCGAGGAGGCGCTACCAGC ATTCGATGGGAATCTGTCGATTCCCGCGGTTGTAGGCGCGCAGCCGCCCTTCTTCCTGCCTGTGTCGCCGAGCGCGCCTG GAGCGATCCCCATTGTCTCGCCATCGCCTATGATTCCAGTTCTGCCCAATCCATCCTCGCCAAAGCTATCAGGACACTGT AAGCTCAACTTCTCGGCTCTAACTCCAGCAATCGATCGAACAGCACAAGATTGTCTCGCTCCCTTGGCATTGTACGTGGG CGAGGTTATCTGCTGCCCACAGTTGCAGACACTCTTTCGTCTAGCTCAAGGGCATCACATTAACACGTCCGGTCGGCTCA CTTTCAATCGTACTGAAGCAAGCTACTGTTTCTCCGACATTAGCAGCCTGCTGGTGAGCAAAGGAGCCAACACCACCGTA TCGGAGATATGTTCCATGGAGCCGAAAAGCTTGACAGGAGGGCAGTGTCCGGTGTACGAGACAAAGGAGCTCTACAGGCT TGTCAACACTTCTAGGCTTCTCCCGGCCTGCAAAGGAGTGGACCCCCTGAAAGAATGCTGCAAGCCGGTTTGCCAGCCAG CGCTCGAAGAGGCCGCGTTACAGCTCGCCTCGAATGGCTCATTTGGATTCGTGAAACAGAGCGGACTTGTTCCAGCCTCG GCGGATGATCAAGTGTTGGAAGACTGCAAGGACGTTGTTCTTGCTTGGGTCGCTGGTCAGCTCAATCTAACCGAGTCTAA CACGGCACTCCGGAATCTATTCAGCTGTAAGGTTAACAAAGCGTGTCCTTTGGTTTTTAGCGACGTATCTGCAGTAGTCG ATGATTGCCATGGACTCTCGCCATCCAACGTTACCTGTTGCAGCTCGCTGCACAGATACATCTCCGAAATGCAGCAGCAG AAGCTGATCACGAACTTGCAAGCGCTAGAGTGTGTGAGTTTACTCGGCTCGATGCTGCAAAAGCGGGGTGTTACAAGCAA TATTTACGATCTATGCGGCATCTATCTCAAAGATTTCAGCCTTCAAGGTGCTGGAGATCAAGGGTGCTTGCTAAATAGCT TGCCGTCTGATGTGATGATCACGAGTAGCTCCGGCATCACCTTCACTTGCGACTTGAATGACAACATCGCTGCTCCATGG ATGCCAATGTCTTCGAGTTCCCTGTGCTCTGGAGGAAGGCCAAACATTTCCTTCCCAAAAATCCCACAATCCGCGGCTCC CCTCACGTCTGGAACTAACTTCGCAAGAGCCAGCTCGATCACTGCGTTTGTAGCTCCGCTCCTGATCCTGGTGATACTAT CAACTTCGATTTGA |