
| Microexon ID | Gm_15:15478971-15478985:+ |
| Species | Glycine max | Coordinates | 15:15478971..15478985 |
| Microexon Cluster ID | MEP43 |
| Size | 15 |
| Phase | 0 |
| Pfam Domain Motif | Unknown |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,15,45 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GARCTTCTYAGWCTAGCWTCAACMGAARTACARMGACARTTYTWTCAGGARGCRAGGATTAAGACTGCTGTWTTCATGGGGTGYAACARWGGWGAAATTGARMTYGGT |
| Logo of Microexon-tag DNA Seq | 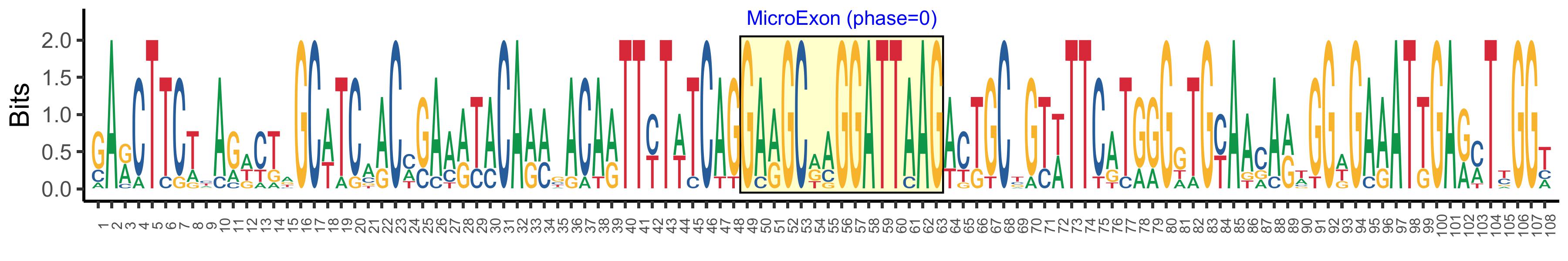 |
| Alignment of exons | 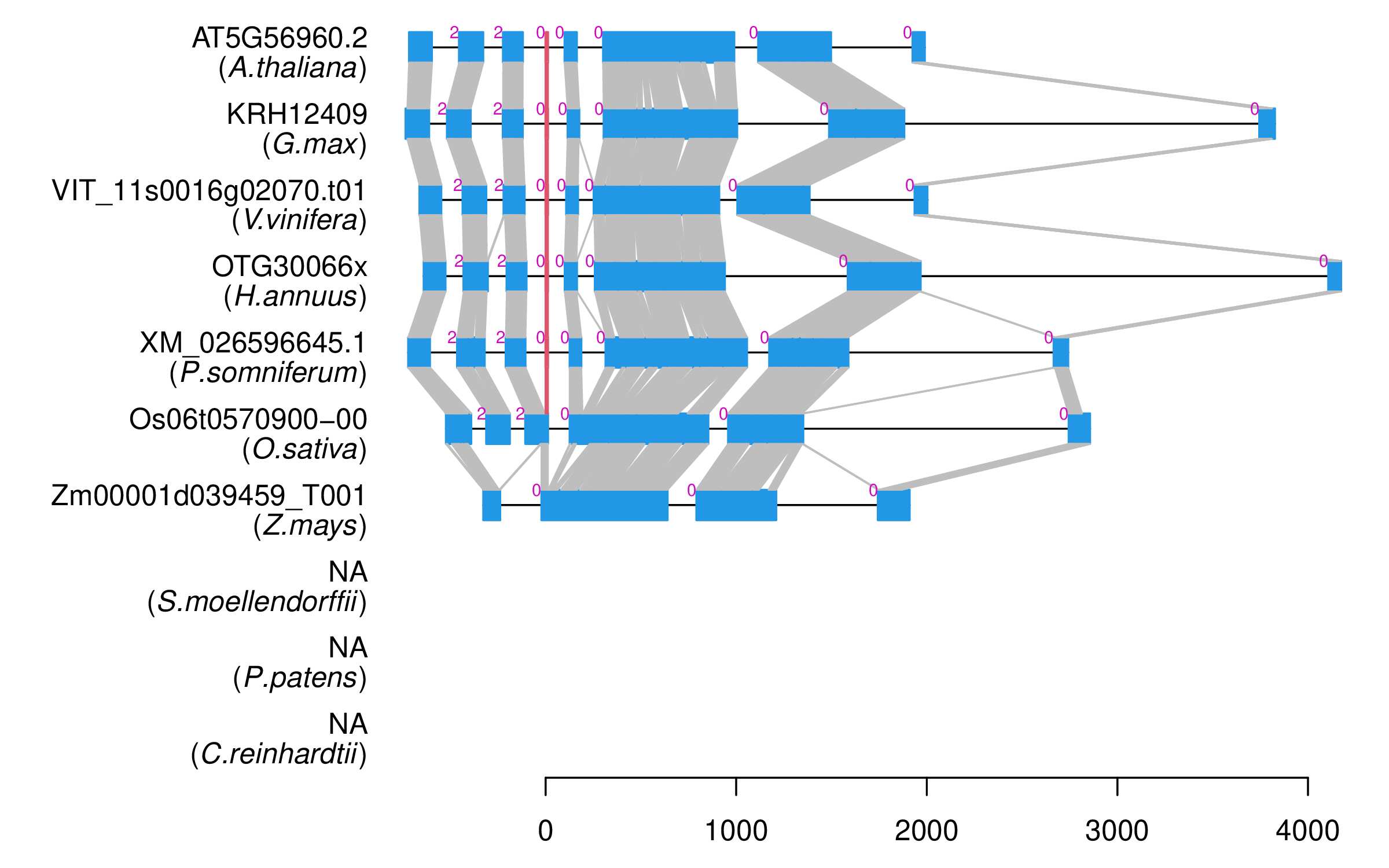 |
| Microexon DNA seq | GAGGCAAGGATTAAG |
| Microexon Amino Acid seq | EARIK |
| Microexon-tag DNA Seq | GAACTTCTCAGATTAGCATCCACAGAAATACAAATACAATTCTTTCAGGAGGCAAGGATTAAGACTGCTGTTTTCATGGGGTGTAACAAGGGAGAAATTGAGCTTGGT |
| Microexon-tag Amino Acid Seq | ELLRLASTEIQIQFFQEARIKTAVFMGCNKGEIELG |
| Microexon-tag spanning region | 15478805-15479131 |
| Microexon-tag prediction score | 0.9797 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRH12409x |
| Reference Transcript ID | KRH12409 |
| Gene ID | GLYMA_15G170500 |
| Gene Name | NA |
| Transcript ID | KRH12409 |
| Protein ID | KRH12409 |
| Gene ID | GLYMA_15G170500 |
| Gene Name | NA |
| Pfam domain motif | Unknown |
| Motif E-value | NA |
| Motif start | NA |
| Motif end | NA |
| Protein seq | >KRH12409 MHPMESVFSLSVAARTDFIQFLVQSLGCSYICLWAYDSISPNRLSFLDGIYNVRNNQASSSLGSVQAQQLFSQFRTLTFD VNDDRVPGLAFRNQRPYLELQQLELLRLASTEIQIQFFQEARIKTAVFMGCNKGEIELGFLNMSQVDIQTALRSLFPEDF STRVQSQQIDQNPPTSSSSSLRSISTGSPEYSSLIFSIPGTSQPHFPPETLGASSSRVEPTMQPVPNSPHQRAIDIQALT EVPSLTQFPTPETEHDAIMRAILHVISPTTSYHHEQQHHQNLPYSNNFLPVVRPDASAFQRYRQDLGSNMASNFRRQSLM KRSLVFFRNMNFMRMRERVQATSRPTNTQLHHMISERRRREKLNENFQALRALLPPGTKKDKASILIAAKETLRSLMAEV DKLSNRNQGLTSLLPAKESTAEETKVASLSPNERLSVRISHVPESSTSEERMVELQVNVRGQVSQTDLLIRLLKFLKLAH HVSLVSMDANTHIAEGNNALHQLTFRLRIIQGSEWDESAFEEAVRRVVADLAQYQMDQ* |
| CDS seq | >KRH12409 ATGCACCCCATGGAAAGTGTCTTCTCTCTCTCTGTAGCCGCCAGAACTGATTTTATCCAATTTCTGGTGCAGTCTTTGGG CTGCAGTTACATTTGTCTATGGGCATATGATTCCATTTCACCAAATCGTTTGAGCTTCTTGGATGGTATCTACAATGTAA GGAACAACCAAGCAAGCTCTAGTTTGGGGAGTGTGCAGGCACAACAGCTTTTCAGTCAGTTCCGGACTTTGACATTTGAT GTCAATGATGACCGTGTTCCTGGACTAGCTTTTAGGAATCAACGACCCTACCTAGAGCTCCAACAATTGGAACTTCTCAG ATTAGCATCCACAGAAATACAAATACAATTCTTTCAGGAGGCAAGGATTAAGACTGCTGTTTTCATGGGGTGTAACAAGG GAGAAATTGAGCTTGGTTTCTTAAATATGTCCCAAGTTGACATTCAAACAGCACTTAGGAGTTTGTTTCCTGAAGATTTT TCCACTAGAGTACAGTCCCAGCAAATTGATCAAAACCCTCCTACATCATCTTCATCTTCTCTGAGATCAATATCCACAGG TAGCCCTGAATACTCATCACTTATATTCAGTATTCCAGGCACATCTCAACCCCACTTTCCACCTGAGACTCTTGGAGCAT CATCATCAAGAGTAGAGCCAACAATGCAGCCAGTGCCAAATAGTCCTCACCAACGAGCCATTGATATTCAAGCATTGACA GAAGTCCCTTCTCTAACCCAATTCCCCACACCAGAAACTGAACATGATGCAATCATGAGAGCAATCCTCCATGTTATATC TCCAACAACTTCTTATCATCATGAGCAGCAGCACCACCAAAACTTGCCTTATAGTAATAATTTTCTTCCTGTGGTGCGTC CTGATGCTAGTGCTTTCCAGAGGTATAGACAAGACTTAGGCTCCAACATGGCCTCAAATTTCAGAAGGCAAAGCCTCATG AAGAGGTCTCTTGTGTTCTTTAGAAACATGAACTTTATGAGAATGAGAGAACGCGTTCAAGCAACGTCTCGTCCCACCAA TACTCAGCTTCATCATATGATATCAGAGAGGAGAAGGCGTGAGAAGCTAAACGAGAATTTTCAAGCACTTAGAGCACTTC TTCCTCCGGGAACAAAGAAAGACAAAGCGTCTATACTAATAGCAGCAAAGGAGACATTGAGGTCTTTGATGGCTGAAGTT GATAAACTAAGCAACAGAAATCAGGGATTGACGTCACTTTTGCCAGCCAAAGAATCTACAGCTGAAGAAACCAAGGTAGC TAGTTTATCACCAAATGAAAGATTAAGTGTCAGAATTTCACATGTACCCGAATCAAGCACATCAGAAGAGAGAATGGTGG AGTTGCAAGTGAATGTGAGAGGACAAGTTTCTCAAACTGATTTATTAATCCGGTTATTGAAATTCTTAAAACTGGCTCAC CACGTGAGTTTGGTTTCCATGGACGCAAACACTCACATTGCAGAAGGGAACAATGCACTTCATCAACTAACCTTCAGATT GAGGATTATTCAGGGAAGTGAATGGGACGAGTCTGCCTTCGAGGAAGCAGTAAGAAGAGTAGTTGCTGACTTGGCACAGT ACCAAATGGACCAATAA |
| Microexon DNA seq | GAGGCAAGGATTAAG |
| Microexon Amino Acid seq | EARIK |
| Microexon-tag DNA Seq | GAACTTCTCAGATTAGCATCCACAGAAATACAAATACAATTCTTTCAGGAGGCAAGGATTAAGACTGCTGTTTTCATGGGGTGTAACAAGGGAGAAATTGAGCTTGGT |
| Microexon-tag Amino Acid seq | ELLRLASTEIQIQFFQEARIKTAVFMGCNKGEIELG |
| Transcript ID | Gm.18257.1 |
| Gene ID | Gm.18257 |
| Gene Name | NA |
| Pfam domain motif | Unknown |
| Motif E-value | NA |
| Motif start | NA |
| Motif end | NA |
| Protein seq | >Gm.18257.1 MLSSKCFFPTRYFSNSLEDQHNYIFATPTCSRLSFLDGIYNVRNNQASSSLGSVQAQQLFSQFRTLTFDVNDDRVPGLAF RNQRPYLELQQLELLRLASTEIQIQFFQEARIKTAVFMGCNKGEIELGFLNMSQVDIQTALRSLFPEDFSTRVQSQQIDQ NPPTSSSSSLRSISTGSPEYSSLIFSIPGTSQPHFPPETLGASSSRVEPTMQPVPNSPHQRAIDIQALTEVPSLTQFPTP ETEHDAIMRAILHVISPTTSYHHEQQHHQNLPYSNNFLPVVRPDASAFQRYRQDLGSNMASNFRRQSLMKRSLVFFRNMN FMRMRERVQATSRPTNTQLHHMISERRRREKLNENFQALRALLPPGTKKDKASILIAAKETLRSLMAEVDKLSNRNQGLT SLLPAKESTAEETKVASLSPNERLSVRISHVPESSTSEERMVELQVNVRGQVSQTDLLIRLLKFLKLAHHVSLVSMDANT HIAEGNNALHQLTFRLRIIQGSEWDESAFEEAVRRVVADLAQYQMDQ* |
| CDS seq | >Gm.18257.1 ATGTTAAGTAGTAAGTGCTTCTTTCCCACTAGGTACTTTTCAAACTCCCTGGAAGACCAACACAATTATATTTTTGCAAC ACCCACATGCAGTCGTTTGAGCTTCTTGGATGGTATCTACAATGTAAGGAACAACCAAGCAAGCTCTAGTTTGGGGAGTG TGCAGGCACAACAGCTTTTCAGTCAGTTCCGGACTTTGACATTTGATGTCAATGATGACCGTGTTCCTGGACTAGCTTTT AGGAATCAACGACCCTACCTAGAGCTCCAACAATTGGAACTTCTCAGATTAGCATCCACAGAAATACAAATACAATTCTT TCAGGAGGCAAGGATTAAGACTGCTGTTTTCATGGGGTGTAACAAGGGAGAAATTGAGCTTGGTTTCTTAAATATGTCCC AAGTTGACATTCAAACAGCACTTAGGAGTTTGTTTCCTGAAGATTTTTCCACTAGAGTACAGTCCCAGCAAATTGATCAA AACCCTCCTACATCATCTTCATCTTCTCTGAGATCAATATCCACAGGTAGCCCTGAATACTCATCACTTATATTCAGTAT TCCAGGCACATCTCAACCCCACTTTCCACCTGAGACTCTTGGAGCATCATCATCAAGAGTAGAGCCAACAATGCAGCCAG TGCCAAATAGTCCTCACCAACGAGCCATTGATATTCAAGCATTGACAGAAGTCCCTTCTCTAACCCAATTCCCCACACCA GAAACTGAACATGATGCAATCATGAGAGCAATCCTCCATGTTATATCTCCAACAACTTCTTATCATCATGAGCAGCAGCA CCACCAAAACTTGCCTTATAGTAATAATTTTCTTCCTGTGGTGCGTCCTGATGCTAGTGCTTTCCAGAGGTATAGACAAG ACTTAGGCTCCAACATGGCCTCAAATTTCAGAAGGCAAAGCCTCATGAAGAGGTCTCTTGTGTTCTTTAGAAACATGAAC TTTATGAGAATGAGAGAACGCGTTCAAGCAACGTCTCGTCCCACCAATACTCAGCTTCATCATATGATATCAGAGAGGAG AAGGCGTGAGAAGCTAAACGAGAATTTTCAAGCACTTAGAGCACTTCTTCCTCCGGGAACAAAGAAAGACAAAGCGTCTA TACTAATAGCAGCAAAGGAGACATTGAGGTCTTTGATGGCTGAAGTTGATAAACTAAGCAACAGAAATCAGGGATTGACG TCACTTTTGCCAGCCAAAGAATCTACAGCTGAAGAAACCAAGGTAGCTAGTTTATCACCAAATGAAAGATTAAGTGTCAG AATTTCACATGTACCCGAATCAAGCACATCAGAAGAGAGAATGGTGGAGTTGCAAGTGAATGTGAGAGGACAAGTTTCTC AAACTGATTTATTAATCCGGTTATTGAAATTCTTAAAACTGGCTCACCACGTGAGTTTGGTTTCCATGGACGCAAACACT CACATTGCAGAAGGGAACAATGCACTTCATCAACTAACCTTCAGATTGAGGATTATTCAGGGAAGTGAATGGGACGAGTC TGCCTTCGAGGAAGCAGTAAGAAGAGTAGTTGCTGACTTGGCACAGTACCAAATGGACCAATAA |