
| Microexon ID | Pp_21:9696233-9696247:+ |
| Species | Physcomitrium patens | Coordinates | 21:9696233..9696247 |
| Microexon Cluster ID | MEP42 |
| Size | 15 |
| Phase | 0 |
| Pfam Domain Motif | bHLH-MYC_N |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,15,45 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GCWMAYKATGCMGAYAGYAAARTYTTYTCTCGMKCTHTKCTWGCMAAGAGTGCWTSWATTCAGACWGTGGTRTGYWTTCCYYWYMTRGRYGGYGTBRTTGARCTWGGY |
| Logo of Microexon-tag DNA Seq | 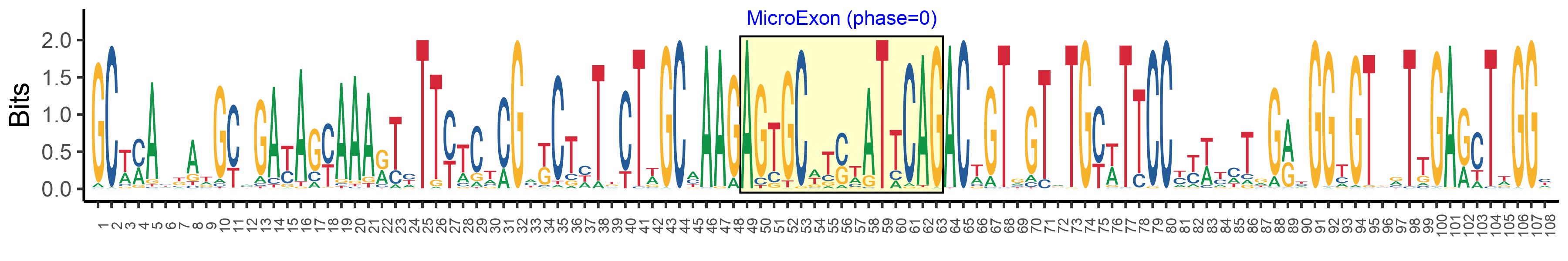 |
| Alignment of exons | 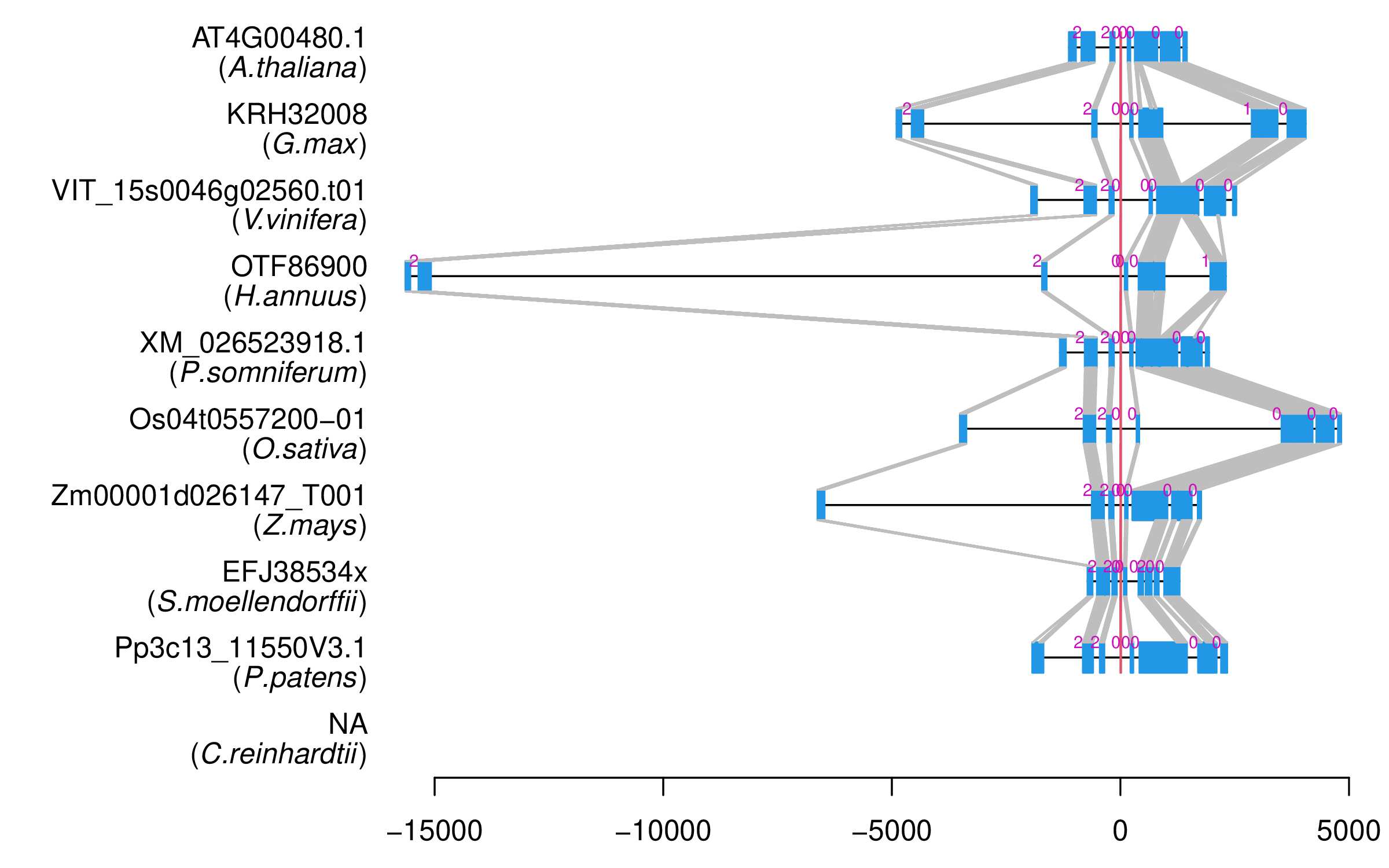 |
| Microexon DNA seq | AGTGCCTCCATTCAG |
| Microexon Amino Acid seq | SASIQ |
| Microexon-tag DNA Seq | ACCAATGAAATATCCACCAAACTTTTTACACGGGCTCTTCTTGCCAAGAGTGCCTCCATTCAGACGATTTTCTGCTTTCCTTTGATGGATGGCGTTGTAGAGTTCGGA |
| Microexon-tag Amino Acid Seq | TNEISTKLFTRALLAKSASIQTIFCFPLMDGVVEFG |
| Microexon-tag spanning region | 9695695-9696424 |
| Microexon-tag prediction score | 0.8911 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | Pp3c21_15150V3.1x |
| Reference Transcript ID | Pp3c21_15150V3.1 |
| Gene ID | Pp3c21_15150 |
| Gene Name | NA |
| Transcript ID | Pp3c21_15150V3.1 |
| Protein ID | Pp3c21_15150V3.1 |
| Gene ID | Pp3c21_15150 |
| Gene Name | NA |
| Pfam domain motif | bHLH-MYC_N |
| Motif E-value | 1.4e-44 |
| Motif start | 20 |
| Motif end | 203 |
| Protein seq | >Pp3c21_15150V3.1 MSGSKLEAIKEMTPLVPPELRLELQAATRAVKWTYSVFWKPASSNQKTLVWGDGYYNGTIKTRKTIGAKELTPEEFGLQR SQQLRDLYNSLSDSKTGHQQASKPFALKPEDLAEQEWFFLLCMSCNFAEGVGLVGRAAADGRYAWQCKTNEISTKLFTRA LLAKSASIQTIFCFPLMDGVVEFGTTEHVPEDSNIVQHVSSFFMERKRHSHPQHSVTTEQKHEHSKNSPQQSENVVHCNQ LAEYGASSKKAKLSSSQMAEVENETSMNRVNSLLDPTSQEQRTYTHLFQQNQGSDSRLYKGENRGKNQSGVLPGRVFSSW KKNSTPSQKSQKAENRQKILKEALFRVTRLYDGAWKNKVDSSFIFTDRAVEDRTSNLGSQKPVPSSEETSASHVLAERRR REKLNDRFVALRELIPNVSKMDKASILGVAIEYVKELQSQLRALESNNTQDGTPRQFGTANEDATITNTTREHLECAGVV HVIDEDKAATSECTITEESFKPGHVNVRVSMNNDVAIVKLHCPYRQTLLVDVLQSLNDLEFDVCGVRSSISDDILSTVLE AKLRSASDGSSPTIIEVEKTLHRAAAGLLKERASASSLQ* |
| CDS seq | >Pp3c21_15150V3.1 ATGTCAGGTTCCAAATTAGAAGCGATAAAGGAAATGACGCCACTTGTGCCACCAGAGTTGCGGCTTGAATTGCAAGCCGC CACTCGTGCTGTTAAATGGACCTACTCCGTCTTCTGGAAGCCAGCGTCCAGTAATCAGAAAACTTTGGTCTGGGGTGATG GCTACTATAATGGAACCATCAAAACTCGAAAAACCATCGGTGCCAAGGAGTTGACGCCTGAGGAGTTCGGCTTGCAGCGT TCGCAGCAGCTCCGTGATCTCTACAACTCATTGTCCGACAGTAAAACTGGGCATCAGCAAGCTAGTAAGCCATTCGCGTT GAAGCCCGAGGATCTTGCAGAACAAGAGTGGTTCTTCCTCCTGTGCATGTCCTGCAATTTCGCTGAGGGTGTAGGGTTGG TAGGAAGAGCAGCAGCAGATGGACGATACGCGTGGCAATGCAAAACCAATGAAATATCCACCAAACTTTTTACACGGGCT CTTCTTGCCAAGAGTGCCTCCATTCAGACGATTTTCTGCTTTCCTTTGATGGATGGCGTTGTAGAGTTCGGAACAACAGA ACATGTACCAGAGGACTCGAATATAGTGCAACATGTTTCGTCTTTTTTCATGGAACGAAAAAGGCATAGCCACCCACAGC ATTCCGTGACCACTGAGCAAAAGCACGAGCATTCGAAGAACAGTCCTCAGCAATCGGAAAATGTGGTTCACTGTAACCAA CTTGCAGAATATGGCGCAAGCTCGAAGAAGGCCAAGCTTTCCAGCTCTCAAATGGCAGAGGTTGAGAATGAAACATCTAT GAACAGAGTAAATAGCCTATTGGATCCGACTAGTCAGGAACAAAGGACATATACACACCTCTTTCAGCAAAACCAGGGCA GTGATTCAAGGCTGTATAAAGGAGAAAATCGGGGGAAAAATCAATCGGGTGTTTTACCTGGGAGAGTATTTAGTTCGTGG AAGAAAAACTCAACGCCTTCTCAGAAATCGCAGAAGGCTGAAAATCGGCAAAAGATTCTGAAAGAGGCTTTGTTTCGTGT CACTCGCCTCTACGATGGTGCGTGGAAGAATAAAGTAGACAGCAGCTTTATATTTACTGACCGCGCTGTTGAGGATAGGA CCTCTAACTTGGGATCTCAAAAACCAGTACCAAGCTCCGAAGAAACATCCGCTAGCCATGTACTAGCTGAGCGTCGAAGG CGGGAGAAACTGAACGACAGATTCGTGGCATTGCGTGAGCTCATTCCTAATGTTTCCAAGATGGATAAAGCATCCATTCT TGGAGTTGCTATTGAATACGTGAAGGAGCTTCAGAGCCAACTCAGAGCTTTGGAGAGTAACAACACGCAGGATGGAACAC CGAGACAATTTGGAACAGCAAATGAGGATGCAACGATCACCAATACCACCAGAGAGCACTTGGAGTGTGCTGGTGTTGTG CACGTAATAGACGAGGACAAGGCAGCAACGTCCGAATGTACGATTACTGAAGAAAGCTTCAAACCTGGGCACGTCAATGT GCGGGTTTCTATGAATAACGATGTAGCTATTGTCAAGCTCCATTGCCCTTACCGACAAACGCTGCTTGTCGACGTGCTGC AGTCACTCAATGATCTCGAGTTTGACGTATGTGGTGTGAGGTCTTCCATATCAGATGACATTCTTTCTACTGTCTTGGAA GCTAAGCTGAGAAGTGCTTCTGATGGCTCAAGTCCAACTATCATCGAGGTGGAAAAGACCTTGCATCGAGCTGCTGCAGG ACTCTTGAAAGAACGAGCCAGTGCTTCTTCACTTCAATGA |
| Microexon DNA seq | AGTGCCTCCATTCAG |
| Microexon Amino Acid seq | SASIQ |
| Microexon-tag DNA Seq | ACCAATGAAATATCCACCAAACTTTTTACACGGGCTCTTCTTGCCAAGAGTGCCTCCATTCAGACGATTTTCTGCTTTCCTTTGATGGATGGCGTTGTAGAGTTCGGA |
| Microexon-tag Amino Acid seq | TNEISTKLFTRALLAKSASIQTIFCFPLMDGVVEFG |
| Transcript ID | Pp.13688.2 |
| Gene ID | Pp.13688 |
| Gene Name | NA |
| Pfam domain motif | bHLH-MYC_N |
| Motif E-value | 2.9e-45 |
| Motif start | 20 |
| Motif end | 203 |
| Protein seq | >Pp.13688.2 MSGSKLEAIKEMTPLVPPELRLELQAATRAVKWTYSVFWKPASSNQKTLVWGDGYYNGTIKTRKTIGAKELTPEEFGLQR SQQLRDLYNSLSDSKTGHQQASKPFALKPEDLAEQEWFFLLCMSCNFAEGVGLVGRAAADGRYAWQCKTNEISTKLFTRA LLAKSASIQTIFCFPLMDGVVEFGTTEHVPEDSNIVQHVSSFFMERKRHSHPQHSVTTEQKHEHSKNSPQQSENVVHCNQ LAEYGASSKKAKLSSSQMAEVENETSMNRVNSLLDPTSQEQRTYTHLFQQNQALWAPSLTSLISPTF* |
| CDS seq | >Pp.13688.2 ATGTCAGGTTCCAAATTAGAAGCGATAAAGGAAATGACGCCACTTGTGCCACCAGAGTTGCGGCTTGAATTGCAAGCCGC CACTCGTGCTGTTAAATGGACCTACTCCGTCTTCTGGAAGCCAGCGTCCAGTAATCAGAAAACTTTGGTCTGGGGTGATG GCTACTATAATGGAACCATCAAAACTCGAAAAACCATCGGTGCCAAGGAGTTGACGCCTGAGGAGTTCGGCTTGCAGCGT TCGCAGCAGCTCCGTGATCTCTACAACTCATTGTCCGACAGTAAAACTGGGCATCAGCAAGCTAGTAAGCCATTCGCGTT GAAGCCCGAGGATCTTGCAGAACAAGAGTGGTTCTTCCTCCTGTGCATGTCCTGCAATTTCGCTGAGGGTGTAGGGTTGG TAGGAAGAGCAGCAGCAGATGGACGATACGCGTGGCAATGCAAAACCAATGAAATATCCACCAAACTTTTTACACGGGCT CTTCTTGCCAAGAGTGCCTCCATTCAGACGATTTTCTGCTTTCCTTTGATGGATGGCGTTGTAGAGTTCGGAACAACAGA ACATGTACCAGAGGACTCGAATATAGTGCAACATGTTTCGTCTTTTTTCATGGAACGAAAAAGGCATAGCCACCCACAGC ATTCCGTGACCACTGAGCAAAAGCACGAGCATTCGAAGAACAGTCCTCAGCAATCGGAAAATGTGGTTCACTGTAACCAA CTTGCAGAATATGGCGCAAGCTCGAAGAAGGCCAAGCTTTCCAGCTCTCAAATGGCAGAGGTTGAGAATGAAACATCTAT GAACAGAGTAAATAGCCTATTGGATCCGACTAGTCAGGAACAAAGGACATATACACACCTCTTTCAGCAAAACCAGGCAC TTTGGGCACCAAGCCTCACGAGCTTGATCTCTCCTACTTTTTGA |