
| Microexon ID | Gm_3:950349-950363:+ |
| Species | Glycine max | Coordinates | 3:950349..950363 |
| Microexon Cluster ID | MEP42 |
| Size | 15 |
| Phase | 0 |
| Pfam Domain Motif | bHLH-MYC_N |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,15,45 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GCWMAYKATGCMGAYAGYAAARTYTTYTCTCGMKCTHTKCTWGCMAAGAGTGCWTSWATTCAGACWGTGGTRTGYWTTCCYYWYMTRGRYGGYGTBRTTGARCTWGGY |
| Logo of Microexon-tag DNA Seq | 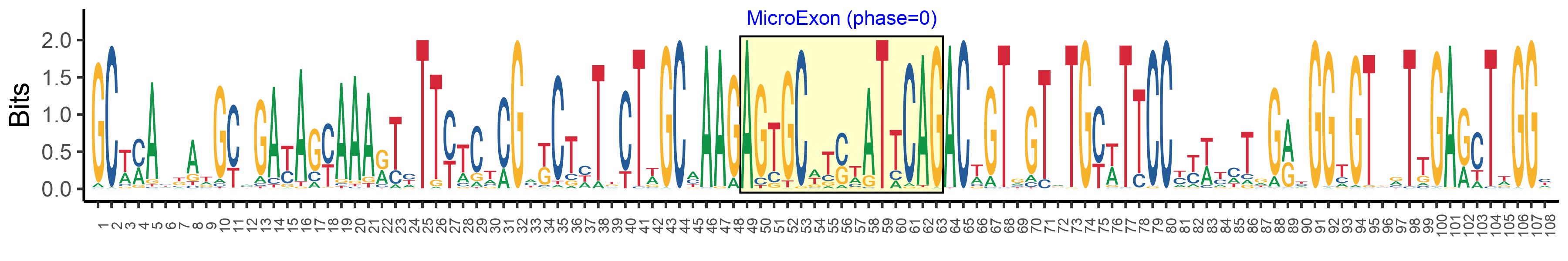 |
| Alignment of exons | 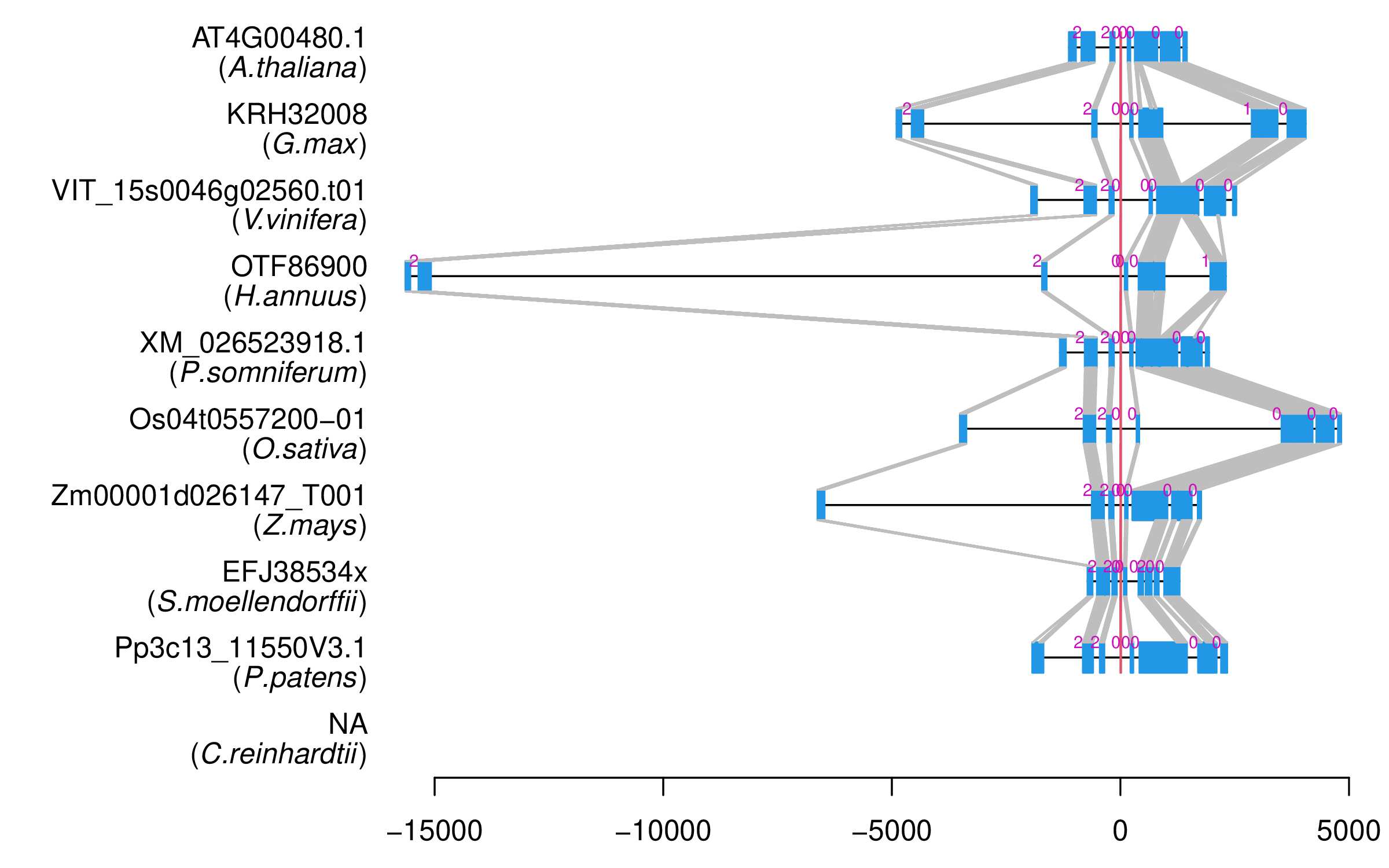 |
| Microexon DNA seq | AGTGCGACAATTCAG |
| Microexon Amino Acid seq | SATIQ |
| Microexon-tag DNA Seq | GCTCAACATGCGGATAGTAAAGTTTTCTCTCGTTCTTTGCTAGCAAAGAGTGCGACAATTCAGACCGTGGTGTGTTTTCCCTATCAGAAAGGCGTTATTGAGATAGGC |
| Microexon-tag Amino Acid Seq | AQHADSKVFSRSLLAKSATIQTVVCFPYQKGVIEIG |
| Microexon-tag spanning region | 949102-950914 |
| Microexon-tag prediction score | 0.9512 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRH65046x |
| Reference Transcript ID | KRH65046 |
| Gene ID | GLYMA_03G009500 |
| Gene Name | NA |
| Transcript ID | KRH65046 |
| Protein ID | KRH65046 |
| Gene ID | GLYMA_03G009500 |
| Gene Name | NA |
| Pfam domain motif | bHLH-MYC_N |
| Motif E-value | 3.8e-48 |
| Motif start | 18 |
| Motif end | 202 |
| Protein seq | >KRH65046 MANNGSPKHEKMQKNLCTQLAVAVRSIQWSYGIFWSPSTTEERVLEWREGYYNGDIKTRKTVQATELEIKADKIGLQRSE QLKELYKFLLAGEADHPQTKRPSVALAPEDLSDLEWYYLVCMSFVFNHNQSLPGRALEIGDTVWLCNAQHADSKVFSRSL LAKSATIQTVVCFPYQKGVIEIGTTELVAEDPSLIQHVKACFLEISKPTCSDKSSSILDKPHDDKYPTCTKGDQRVLDTM ALENPCSLEEKIKFDHEPINELQDDNNEGSNGCEHHFPMDGSMIEGINGVPSQVHFVNDDALVIGAPDSLSSCDCMSEAS ENQGKDSKNVGQTQLMELQDCHKPKRSSLDVGADEDLCYIRTLCAILGNSSTFKPNPYAGNSNCKSSFAKWKKGRVSERK RPKLHQSMLKKTLFKVPFMHRSYSSLKSQKGNDRMEWTSKLENDDHGLIGKAFSDKKREIKNFQVVKSMVPSSISEVEKI SILGDTIKYLKKLETRVEELESYMEVTGPEARKRSKCPDVLEQMSDNYGTRKICMGMKPWMNKRKACGIDEIDTELERIT SEEAKALDVKVNVKDQEVLIEMKCPYRKYILYDIMDTINNLHLDAQTVESSTSDGVLTLTLKSKFRGAATAPMRMIKEAL WKVSGNI* |
| CDS seq | >KRH65046 ATGGCTAACAACGGGAGTCCAAAGCATGAGAAGATGCAAAAGAACCTATGCACACAGCTAGCTGTTGCAGTGAGAAGCAT TCAATGGAGCTATGGTATTTTCTGGTCACCTTCAACCACTGAAGAAAGGGTATTGGAATGGAGGGAAGGCTACTACAATG GAGACATTAAAACAAGAAAGACAGTGCAAGCCACGGAATTGGAAATAAAGGCTGATAAAATAGGCCTGCAGAGGAGTGAG CAACTGAAGGAACTGTACAAGTTTCTTCTTGCAGGTGAAGCTGATCATCCACAAACTAAAAGGCCTTCTGTTGCATTAGC TCCAGAGGACCTCTCAGATTTGGAGTGGTATTACTTGGTTTGCATGTCCTTTGTTTTCAATCACAACCAAAGTTTGCCTG GAAGAGCACTAGAAATTGGTGACACAGTCTGGTTATGCAATGCTCAACATGCGGATAGTAAAGTTTTCTCTCGTTCTTTG CTAGCAAAGAGTGCGACAATTCAGACCGTGGTGTGTTTTCCCTATCAGAAAGGCGTTATTGAGATAGGCACAACTGAACT GGTTGCTGAGGATCCTAGTCTCATTCAACATGTCAAGGCTTGCTTCTTAGAAATCTCAAAGCCTACATGCTCTGATAAAT CTTCCTCTATCCTTGATAAGCCACATGATGACAAATATCCAACATGCACCAAGGGTGACCAAAGGGTGTTAGATACAATG GCTTTAGAGAACCCATGTTCTCTTGAAGAAAAAATCAAATTTGATCATGAACCCATCAATGAATTACAAGATGATAACAA TGAAGGTTCTAATGGTTGTGAGCACCATTTCCCTATGGATGGATCCATGATTGAAGGTATCAATGGTGTGCCATCTCAAG TGCATTTTGTGAATGATGATGCCTTGGTCATTGGTGCTCCTGATTCCCTTAGTTCTTGTGATTGTATGTCTGAAGCTTCT GAGAATCAAGGCAAGGATTCCAAGAATGTAGGCCAAACTCAACTTATGGAACTTCAAGATTGCCATAAACCAAAGAGAAG CTCCTTGGATGTGGGAGCTGATGAGGACTTGTGCTACATAAGAACACTTTGTGCTATTTTGGGGAACTCATCAACTTTTA AGCCAAACCCTTATGCTGGTAACTCAAATTGCAAATCTAGTTTTGCAAAATGGAAGAAAGGGAGAGTTTCTGAAAGGAAG AGGCCAAAATTGCACCAAAGCATGTTAAAGAAGACTTTGTTTAAGGTCCCCTTTATGCACAGAAGTTACTCTTCTCTCAA GTCACAAAAAGGGAATGACAGAATGGAGTGGACTAGTAAATTGGAAAATGATGATCATGGTTTAATAGGGAAAGCATTCT CTGATAAGAAAAGAGAAATTAAAAACTTTCAAGTTGTCAAATCAATGGTTCCATCTTCTATAAGTGAGGTGGAGAAGATT TCAATTCTTGGGGACACAATTAAGTACTTGAAAAAGCTTGAGACAAGAGTGGAAGAGCTAGAATCATACATGGAAGTTAC AGGTCCTGAAGCAAGAAAGAGGAGTAAATGCCCTGATGTTCTAGAGCAGATGTCAGATAACTATGGCACCAGAAAGATTT GCATGGGAATGAAGCCTTGGATGAACAAGAGGAAGGCTTGTGGTATTGATGAGATAGACACAGAGCTAGAAAGAATTACT TCTGAAGAAGCAAAAGCTTTGGATGTGAAAGTCAATGTGAAGGATCAGGAGGTTCTGATTGAGATGAAATGTCCTTACAG GAAATACATACTGTATGATATCATGGATACCATTAACAACCTACATTTAGATGCTCAGACAGTTGAATCTTCAACAAGTG ATGGTGTTCTCACATTGACACTTAAATCTAAGTTTCGAGGAGCAGCAACAGCACCAATGAGGATGATCAAAGAAGCGCTG TGGAAAGTATCTGGAAATATTTGA |
| Microexon DNA seq | AGTGCGACAATTCAG |
| Microexon Amino Acid seq | SATIQ |
| Microexon-tag DNA Seq | GCTCAACATGCGGATAGTAAAGTTTTCTCTCGTTCTTTGCTAGCAAAGAGTGCGACAATTCAGACCGTGGTGTGTTTTCCCTATCAGAAAGGCGTTATTGAGATAGGC |
| Microexon-tag Amino Acid seq | AQHADSKVFSRSLLAKSATIQTVVCFPYQKGVIEIG |
| Transcript ID | Gm.34933.1 |
| Gene ID | Gm.34933 |
| Gene Name | NA |
| Pfam domain motif | bHLH-MYC_N |
| Motif E-value | 2.6e-43 |
| Motif start | 18 |
| Motif end | 194 |
| Protein seq | >Gm.34933.1 MANNGSPKHEKMQKNLCTQLAVAVRSIQWSYGIFWSPSTTEERVLEWREGYYNGDIKTRKTVQATELEIKADKIGLQRSE QLKELYKFLLAGEADHPQTKRPSVALAPEDLSDLEWYYLVCMSFVFNHNQSLPGRALEIGDTVWLCNAQHADSKVFSRSL LAKSATIQTVVCFPYQKGVIEIGTTELVEKISILGDTIKYLKKLETRVEELESYMEVTGPEARKRSKCPDVLEQMSDNYG TRKICMGMKPWMNKRKACGIDEIDTELERITSEEAKALDVKVNVKDQEVLIEMKCPYRKYILYDIMDTINNLHLDAQTVE SSTSDGVLTLTLKSKFRGAATAPMRMIKEALWKVSGNI* |
| CDS seq | >Gm.34933.1 ATGGCTAACAACGGGAGTCCAAAGCATGAGAAGATGCAAAAGAACCTATGCACACAGCTAGCTGTTGCAGTGAGAAGCAT TCAATGGAGCTATGGTATTTTCTGGTCACCTTCAACCACTGAAGAAAGGGTATTGGAATGGAGGGAAGGCTACTACAATG GAGACATTAAAACAAGAAAGACAGTGCAAGCCACGGAATTGGAAATAAAGGCTGATAAAATAGGCCTGCAGAGGAGTGAG CAACTGAAGGAACTGTACAAGTTTCTTCTTGCAGGTGAAGCTGATCATCCACAAACTAAAAGGCCTTCTGTTGCATTAGC TCCAGAGGACCTCTCAGATTTGGAGTGGTATTACTTGGTTTGCATGTCCTTTGTTTTCAATCACAACCAAAGTTTGCCTG GAAGAGCACTAGAAATTGGTGACACAGTCTGGTTATGCAATGCTCAACATGCGGATAGTAAAGTTTTCTCTCGTTCTTTG CTAGCAAAGAGTGCGACAATTCAGACCGTGGTGTGTTTTCCCTATCAGAAAGGCGTTATTGAGATAGGCACAACTGAACT GGTGGAGAAGATTTCAATTCTTGGGGACACAATTAAGTACTTGAAAAAGCTTGAGACAAGAGTGGAAGAGCTAGAATCAT ACATGGAAGTTACAGGTCCTGAAGCAAGAAAGAGGAGTAAATGCCCTGATGTTCTAGAGCAGATGTCAGATAACTATGGC ACCAGAAAGATTTGCATGGGAATGAAGCCTTGGATGAACAAGAGGAAGGCTTGTGGTATTGATGAGATAGACACAGAGCT AGAAAGAATTACTTCTGAAGAAGCAAAAGCTTTGGATGTGAAAGTCAATGTGAAGGATCAGGAGGTTCTGATTGAGATGA AATGTCCTTACAGGAAATACATACTGTATGATATCATGGATACCATTAACAACCTACATTTAGATGCTCAGACAGTTGAA TCTTCAACAAGTGATGGTGTTCTCACATTGACACTTAAATCTAAGTTTCGAGGAGCAGCAACAGCACCAATGAGGATGAT CAAAGAAGCGCTGTGGAAAGTATCTGGAAATATTTGA |