
| Microexon ID | Gm_2:15256475-15256489:- |
| Species | Glycine max | Coordinates | 2:15256475..15256489 |
| Microexon Cluster ID | MEP42 |
| Size | 15 |
| Phase | 0 |
| Pfam Domain Motif | bHLH-MYC_N |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,15,45 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GCWMAYKATGCMGAYAGYAAARTYTTYTCTCGMKCTHTKCTWGCMAAGAGTGCWTSWATTCAGACWGTGGTRTGYWTTCCYYWYMTRGRYGGYGTBRTTGARCTWGGY |
| Logo of Microexon-tag DNA Seq | 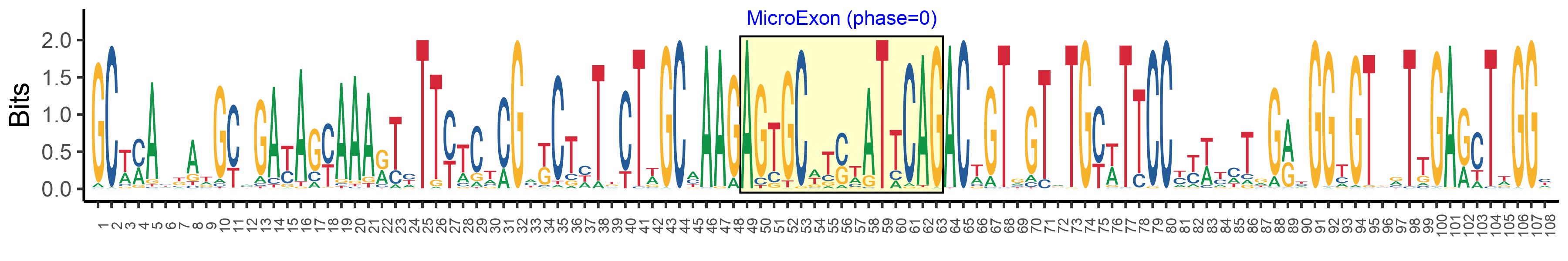 |
| Alignment of exons | 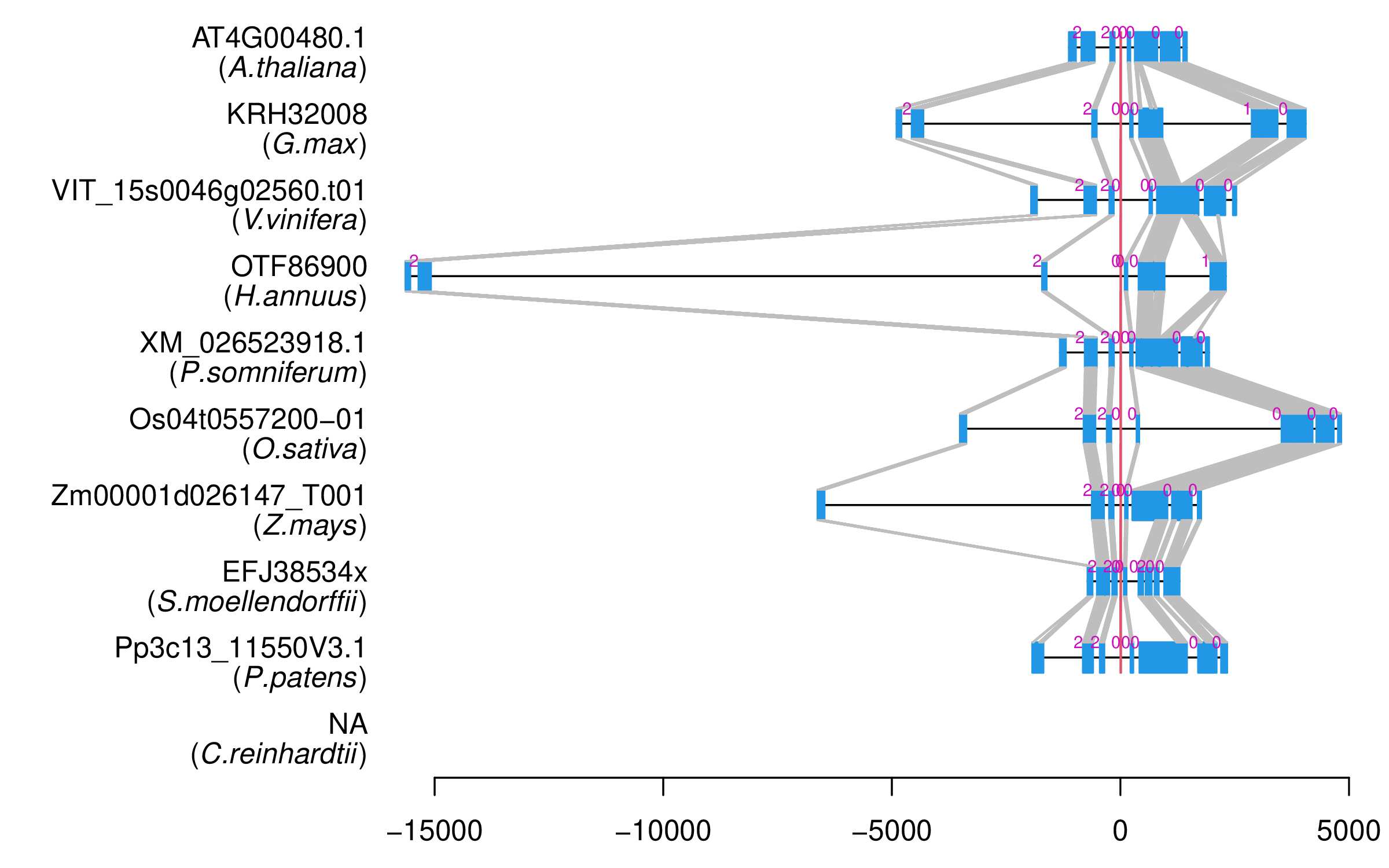 |
| Microexon DNA seq | AGTGCTCGTATACAG |
| Microexon Amino Acid seq | SARIQ |
| Microexon-tag DNA Seq | GCAAACGAGGTGGACAGCAAAACATTTTCACGAGCTATTCTAGCCAAGAGTGCTCGTATACAGACCGTGGTGTGCATTCCTTTATTGGATGGCGTCGTTGAATTTGGG |
| Microexon-tag Amino Acid Seq | ANEVDSKTFSRAILAKSARIQTVVCIPLLDGVVEFG |
| Microexon-tag spanning region | 15256260-15256826 |
| Microexon-tag prediction score | 0.9472 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRH71432x |
| Reference Transcript ID | KRH71432 |
| Gene ID | GLYMA_02G147800 |
| Gene Name | NA |
| Transcript ID | KRH71432 |
| Protein ID | KRH71432 |
| Gene ID | GLYMA_02G147800 |
| Gene Name | NA |
| Pfam domain motif | bHLH-MYC_N |
| Motif E-value | 9.2e-53 |
| Motif start | 9 |
| Motif end | 191 |
| Protein seq | >KRH71432 MTAPLDTGLQSMLQAAVQSVHWTYSLFWQLCPQQVILVWGDGYYNGAIKTRKTVQPMEVSAEEASLQRSQQLRELYESLS VGETNPPTRRPCAALSPEDLTESEWFYLMCVSFSFPPGVGLPGKAYARRQHLWLTGANEVDSKTFSRAILAKSARIQTVV CIPLLDGVVEFGTTDKVQEDLSFIQHVKTFFIDHLIPLRPKPALSEHSTSNPTSSDHIPTVMYTMVDPPAAKCNLNDDMD EDIEEEEEEEEEDEVESGSEDETGDGIACQTLRPSTVAEPSELMQLEMPEDIRLGSPNDGSNNLDSDFHLLAVSQGGNEA RQAESTRRWSSSQEPMQVQLPTSALHPLEDLTQEDTHYSQTVSNILQNQTTRWLASPSSIGYNTYSTHSAFAKWSSRASH HFHPAADGTSQWLLKYILFTVPHLHAKNPGESSPHTAADTKLRGKGTPQDELSANHVLAERRRREKLNERFIILRSLVPF VTKMDKASILGDTIEYVKQLRRKIQELEARNRLTEERSKLPEVAVQRTSSSSSKEQQRSGVTMMEKRKVRIVEGVAAKAK AVEVEATTSVQVSIIESDALLEIECRHREGLLLDVMQMLREVRIEVIGVQSSLNNGVFVAELRAKVKEHANGKKVSIVEV KRALNQIIPHAVD* |
| CDS seq | >KRH71432 ATGACTGCGCCACTAGACACTGGCCTTCAAAGCATGTTGCAGGCTGCGGTGCAATCTGTTCACTGGACTTACAGCCTCTT CTGGCAACTTTGTCCACAGCAAGTGATTCTGGTTTGGGGTGATGGTTACTACAATGGAGCAATTAAGACACGGAAGACGG TGCAACCAATGGAGGTGAGTGCTGAGGAGGCTTCTCTCCAAAGAAGTCAGCAACTTAGAGAACTTTATGAATCATTGTCC GTCGGAGAGACAAATCCGCCGACTCGCCGGCCTTGTGCTGCCTTGTCGCCGGAGGACTTAACAGAATCTGAATGGTTCTA TTTGATGTGTGTCTCATTCTCCTTTCCTCCTGGTGTTGGGTTGCCTGGAAAGGCATATGCCAGGAGGCAGCATCTATGGC TCACTGGTGCAAACGAGGTGGACAGCAAAACATTTTCACGAGCTATTCTAGCCAAGAGTGCTCGTATACAGACCGTGGTG TGCATTCCTTTATTGGATGGCGTCGTTGAATTTGGGACAACAGATAAGGTTCAAGAAGATCTTAGCTTCATCCAACACGT GAAGACTTTTTTCATAGACCATCTCATCCCTCTCAGGCCAAAGCCTGCCCTATCGGAGCACTCAACCTCCAACCCTACCT CCTCCGACCACATCCCCACAGTCATGTACACCATGGTGGACCCACCTGCCGCCAAGTGCAACCTAAATGATGACATGGAT GAGGATATTGAAGAGGAAGAGGAGGAGGAAGAAGAGGACGAGGTGGAATCCGGTTCCGAAGATGAAACTGGAGACGGTAT TGCCTGCCAAACTTTGAGACCATCAACGGTGGCAGAGCCCAGCGAGCTGATGCAGCTGGAGATGCCAGAGGACATTCGAC TCGGGTCGCCGAACGATGGATCAAACAACCTGGACTCAGACTTTCATTTGCTTGCGGTGAGCCAGGGTGGCAATGAGGCA AGGCAGGCTGAGTCAACTCGAAGGTGGAGCTCAAGTCAAGAACCCATGCAAGTTCAACTACCGACTTCAGCCCTTCATCC ATTAGAAGACTTGACCCAAGAGGACACTCACTACTCTCAAACGGTGTCCAACATCCTCCAAAACCAGACCACACGGTGGC TCGCCTCACCCTCCTCCATTGGCTACAACACATACTCCACGCATTCGGCATTCGCAAAATGGAGCAGCCGCGCCAGCCAC CACTTCCACCCGGCGGCGGACGGCACCAGCCAGTGGCTCCTCAAGTACATCCTCTTCACGGTCCCGCACCTCCACGCCAA GAACCCCGGCGAGAGTTCTCCCCATACCGCGGCGGACACCAAGCTCCGCGGCAAGGGGACGCCGCAGGACGAGCTCAGTG CGAACCACGTGCTGGCCGAGCGCCGCCGCCGCGAGAAGCTGAATGAGAGGTTCATAATCCTGCGGTCGCTGGTCCCCTTC GTGACCAAAATGGACAAGGCGTCGATATTGGGAGACACCATCGAGTACGTGAAGCAGTTGCGGCGGAAGATCCAGGAGCT CGAGGCGCGTAACCGCTTAACGGAGGAGCGTTCCAAGCTCCCAGAGGTAGCAGTTCAGAGGACGAGTAGCAGTTCGAGCA AGGAGCAACAGAGGAGTGGGGTGACGATGATGGAAAAGAGGAAGGTGAGGATCGTGGAAGGGGTAGCGGCGAAGGCAAAG GCAGTGGAAGTTGAGGCCACCACGTCGGTGCAAGTTTCGATCATAGAGAGCGACGCGCTGTTGGAGATTGAGTGTCGTCA CAGAGAAGGGTTACTTCTGGACGTGATGCAGATGCTGAGGGAGGTGAGGATAGAGGTTATAGGGGTGCAGTCTTCGCTTA ACAATGGGGTGTTCGTGGCGGAGTTGAGGGCTAAGGTGAAGGAGCATGCAAACGGGAAGAAGGTCAGCATTGTGGAAGTG AAGAGGGCACTTAACCAAATTATACCCCATGCCGTTGACTAA |
| Microexon DNA seq | AGTGCTCGTATACAG |
| Microexon Amino Acid seq | SARIQ |
| Microexon-tag DNA Seq | GCAAACGAGGTGGACAGCAAAACATTTTCACGAGCTATTCTAGCCAAGAGTGCTCGTATACAGACCGTGGTGTGCATTCCTTTATTGGATGGCGTCGTTGAATTTGGG |
| Microexon-tag Amino Acid seq | ANEVDSKTFSRAILAKSARIQTVVCIPLLDGVVEFG |
| Transcript ID | Gm.30884.1 |
| Gene ID | Gm.30884 |
| Gene Name | NA |
| Pfam domain motif | bHLH-MYC_N |
| Motif E-value | 2.6e-53 |
| Motif start | 9 |
| Motif end | 191 |
| Protein seq | >Gm.30884.1 MTAPLDTGLQSMLQAAVQSVHWTYSLFWQLCPQQVILVWGDGYYNGAIKTRKTVQPMEVSAEEASLQRSQQLRELYESLS VGETNPPTRRPCAALSPEDLTESEWFYLMCVSFSFPPGVGLPGKAYARRQHLWLTGANEVDSKTFSRAILAKSARIQTVV CIPLLDGVVEFGTTDKVQEDLSFIQHVKTFFIDHLIPLRPKPALSEHSTSNPTSSDHIPTVMYTMVDPPAAKCNLNDDMD EDIEEEEEEEEEDEVESGSEDETGDGIACQTLRPSTVAEPSELMQLEMPEDIRLGSPNDGSNNLDSDFHLLAVSQGGNEA RQAESTRRWSSSQEPMQVQLPTSGHVVHPNKFEVLDHLVGVLI* |
| CDS seq | >Gm.30884.1 ATGACTGCGCCACTAGACACTGGCCTTCAAAGCATGTTGCAGGCTGCGGTGCAATCTGTTCACTGGACTTACAGCCTCTT CTGGCAACTTTGTCCACAGCAAGTGATTCTGGTTTGGGGTGATGGTTACTACAATGGAGCAATTAAGACACGGAAGACGG TGCAACCAATGGAGGTGAGTGCTGAGGAGGCTTCTCTCCAAAGAAGTCAGCAACTTAGAGAACTTTATGAATCATTGTCC GTCGGAGAGACAAATCCGCCGACTCGCCGGCCTTGTGCTGCCTTGTCGCCGGAGGACTTAACAGAATCTGAATGGTTCTA TTTGATGTGTGTCTCATTCTCCTTTCCTCCTGGTGTTGGGTTGCCTGGAAAGGCATATGCCAGGAGGCAGCATCTATGGC TCACTGGTGCAAACGAGGTGGACAGCAAAACATTTTCACGAGCTATTCTAGCCAAGAGTGCTCGTATACAGACCGTGGTG TGCATTCCTTTATTGGATGGCGTCGTTGAATTTGGGACAACAGATAAGGTTCAAGAAGATCTTAGCTTCATCCAACACGT GAAGACTTTTTTCATAGACCATCTCATCCCTCTCAGGCCAAAGCCTGCCCTATCGGAGCACTCAACCTCCAACCCTACCT CCTCCGACCACATCCCCACAGTCATGTACACCATGGTGGACCCACCTGCCGCCAAGTGCAACCTAAATGATGACATGGAT GAGGATATTGAAGAGGAAGAGGAGGAGGAAGAAGAGGACGAGGTGGAATCCGGTTCCGAAGATGAAACTGGAGACGGTAT TGCCTGCCAAACTTTGAGACCATCAACGGTGGCAGAGCCCAGCGAGCTGATGCAGCTGGAGATGCCAGAGGACATTCGAC TCGGGTCGCCGAACGATGGATCAAACAACCTGGACTCAGACTTTCATTTGCTTGCGGTGAGCCAGGGTGGCAATGAGGCA AGGCAGGCTGAGTCAACTCGAAGGTGGAGCTCAAGTCAAGAACCCATGCAAGTTCAACTACCGACTTCAGGGCATGTGGT CCATCCTAATAAATTTGAGGTCTTGGACCATTTGGTGGGGGTCCTCATCTAA |