
| Microexon ID | Gm_19:41561718-41561732:+ |
| Species | Glycine max | Coordinates | 19:41561718..41561732 |
| Microexon Cluster ID | MEP42 |
| Size | 15 |
| Phase | 0 |
| Pfam Domain Motif | bHLH-MYC_N |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,15,45 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GCWMAYKATGCMGAYAGYAAARTYTTYTCTCGMKCTHTKCTWGCMAAGAGTGCWTSWATTCAGACWGTGGTRTGYWTTCCYYWYMTRGRYGGYGTBRTTGARCTWGGY |
| Logo of Microexon-tag DNA Seq | 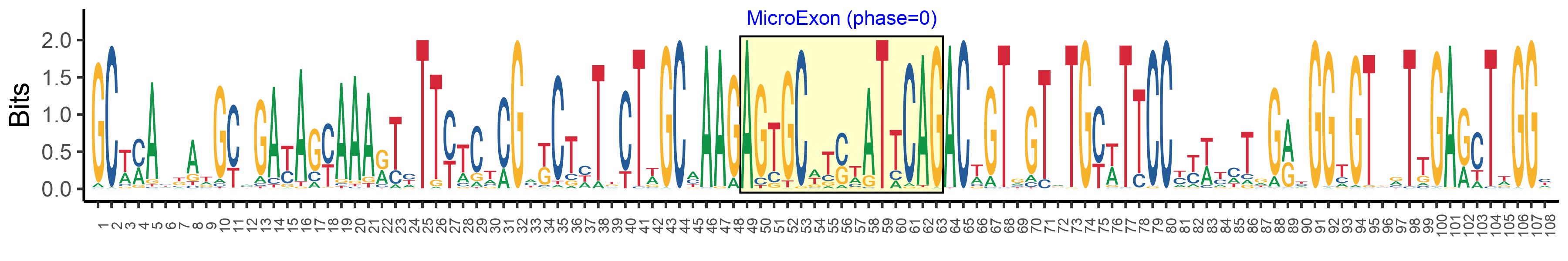 |
| Alignment of exons | 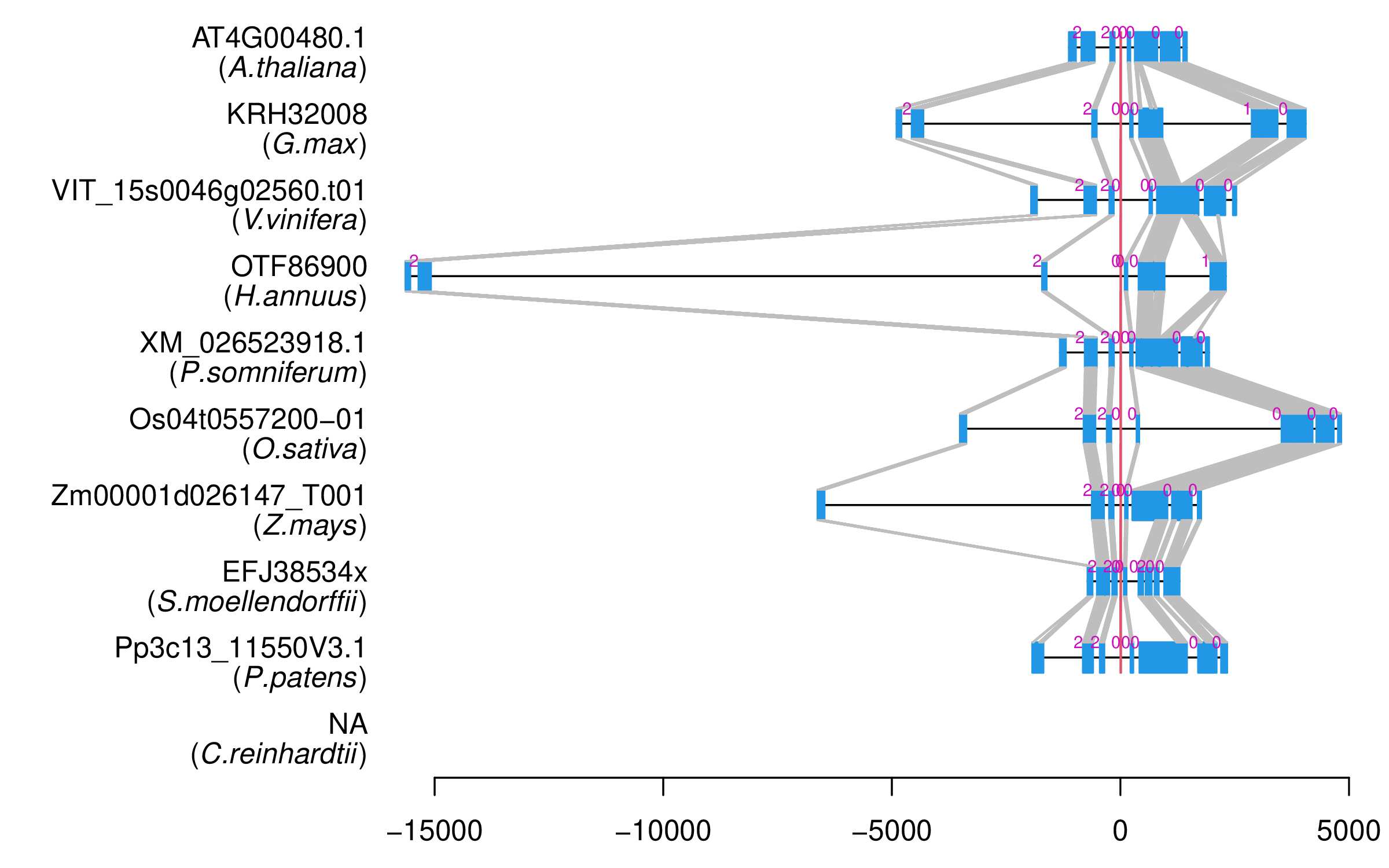 |
| Microexon DNA seq | AGTGCTCATATACAG |
| Microexon Amino Acid seq | SAHIQ |
| Microexon-tag DNA Seq | GCAAACGAAGTAGACAGCAAAACTTTTTCAAGAGCTATTCTTGCCAAGAGTGCTCATATACAGACAGTGGTGTGTATTCCTGTGTTGGAGGGCGTCGTTGAGTTGGGC |
| Microexon-tag Amino Acid Seq | ANEVDSKTFSRAILAKSAHIQTVVCIPVLEGVVELG |
| Microexon-tag spanning region | 41561310-41562706 |
| Microexon-tag prediction score | 0.9375 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRG95511x |
| Reference Transcript ID | KRG95511 |
| Gene ID | GLYMA_19G155300 |
| Gene Name | NA |
| Transcript ID | KRG95511 |
| Protein ID | KRG95511 |
| Gene ID | GLYMA_19G155300 |
| Gene Name | NA |
| Pfam domain motif | bHLH-MYC_N |
| Motif E-value | 1.1e-52 |
| Motif start | 9 |
| Motif end | 192 |
| Protein seq | >KRG95511 MATPPNSRLHTMLRASVQSVQWTYSLFWQLCPQQGILTWGDGYYNGAIKTRKTVQAMEVSTEEASLQRSEQLRELYESLS AEETINTQTRRPCAALSPEDLTESEWFYLLCVSFSFHLGIGLPGTAYARRQHLWLSGANEVDSKTFSRAILAKSAHIQTV VCIPVLEGVVELGTTDKIEEDLNFIQHIKSFFIDQQPPPPTAKPALSEHSTSNLTSSYPLVIVPVTAAATANNVLIQNDM NIVDKGEAIILNNNNNNTEAELLADPNSNSFIPSELMELDHQLEEFGVGSPGDGSNHLDSFPKEESMALCAAGLELLQLQ RPPAPAHPPTENLAQGDTDTHYSQTVSSILKKNSSRWWPDSPSVNHPTDSFQSAFNKWKSDTDNHHHYFHETVADGTSQG LLKYILFNVPYLHANRLKGTGASSYETNHVMAERRRREKLNERFLILRSMVPFMMRMDKESILEDTIHYIKQLREKIESL EARERLRGKRRVREVEVSIIESEALLEVECVHRERLLLDVMTMLRELGVEVMMVQSWVKDDGVFVAEMRAKVKENGNGKK ASVVEVKNALNQIIPHHEPYTLCSNDHF* |
| CDS seq | >KRG95511 ATGGCTACGCCGCCAAATAGCAGGCTTCACACCATGTTGCGGGCATCAGTGCAATCAGTTCAGTGGACTTACAGCCTCTT CTGGCAACTTTGTCCACAACAAGGGATATTGACTTGGGGTGATGGATACTACAACGGAGCAATTAAGACACGGAAGACAG TGCAAGCAATGGAGGTAAGCACTGAGGAAGCGTCTCTGCAAAGAAGCGAGCAACTAAGGGAGTTATATGAATCTTTGTCT GCTGAGGAGACAATAAACACGCAAACACGAAGGCCATGTGCTGCTTTGTCGCCTGAGGATCTAACTGAATCCGAATGGTT CTATTTGTTGTGTGTCTCTTTCTCCTTTCATCTTGGTATCGGGTTGCCCGGAACAGCATATGCCAGGAGGCAGCATTTAT GGCTCAGTGGTGCAAACGAAGTAGACAGCAAAACTTTTTCAAGAGCTATTCTTGCCAAGAGTGCTCATATACAGACAGTG GTGTGTATTCCTGTGTTGGAGGGCGTCGTTGAGTTGGGCACAACCGATAAGATCGAAGAAGACCTAAATTTCATCCAACA CATAAAGAGCTTCTTCATAGATCAGCAGCCTCCTCCTCCCACGGCAAAGCCTGCACTGTCAGAGCACTCCACCTCCAATC TCACTTCTTCATACCCTCTTGTTATTGTTCCTGTCACTGCTGCCGCCACCGCCAATAATGTTCTCATCCAAAACGACATG AACATTGTTGATAAAGGAGAAGCTATTATATTGAATAATAATAATAATAATACCGAAGCAGAATTATTAGCAGACCCTAA CAGTAACAGTTTCATACCGAGCGAGCTAATGGAGCTGGACCACCAGTTGGAGGAATTCGGGGTCGGATCACCTGGAGATG GCTCCAATCACTTGGATTCCTTCCCCAAAGAGGAGTCAATGGCACTGTGTGCTGCAGGGTTGGAGCTCCTTCAACTTCAA CGACCACCAGCTCCAGCACATCCTCCCACAGAAAATCTAGCACAAGGCGACACAGACACCCACTACTCTCAAACAGTGTC CAGCATCCTTAAAAAGAACTCCAGCAGGTGGTGGCCGGATTCACCCTCCGTCAACCACCCCACTGACTCCTTCCAATCAG CTTTCAACAAATGGAAATCCGACACTGACAACCACCACCACTATTTCCACGAGACGGTGGCCGATGGCACCTCGCAGGGG CTCCTCAAGTACATTTTGTTCAACGTCCCTTACTTGCACGCCAACCGGCTCAAGGGCACGGGAGCTTCGTCATACGAGAC TAATCACGTGATGGCGGAGCGCCGCAGAAGGGAGAAGTTGAATGAGAGGTTTCTCATTCTACGTTCGATGGTACCTTTCA TGATGAGGATGGACAAGGAGTCCATTCTGGAAGATACCATCCACTACATCAAACAGTTAAGAGAGAAGATTGAGAGTCTC GAAGCGCGTGAGCGCCTCCGGGGGAAGAGGAGAGTGAGGGAGGTGGAGGTTTCGATCATAGAGAGCGAGGCGCTGTTGGA GGTGGAGTGTGTGCATAGGGAAAGGTTGTTGCTGGATGTGATGACAATGTTGAGGGAGTTAGGGGTGGAAGTGATGATGG TGCAGTCTTGGGTTAAGGATGATGGAGTGTTTGTGGCGGAGATGAGGGCCAAGGTGAAGGAAAATGGTAACGGGAAAAAG GCTAGTGTTGTTGAAGTGAAAAATGCCCTTAACCAGATTATACCACATCATGAGCCATACACACTTTGTTCCAATGATCA CTTTTAA |
Gm_19:41561718-41561732:+ does not have available information here.