
| Microexon ID | Pt_4:3240958-3240972:+ |
| Species | Populus Trichocarpa | Coordinates | 4:3240958..3240972 |
| Microexon Cluster ID | MEP41 |
| Size | 15 |
| Phase | 0 |
| Pfam Domain Motif | DUF974 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,15,45 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | CARTTYTTCAAGTTYATTGTTKCWAAYCCACTTTCWGTTAGRACAAAGGTYCGYRYTRTCAAGGAAACTACMTWTYTRGARGCTTGYATWGARAAYCATACAAAATCA |
| Logo of Microexon-tag DNA Seq | 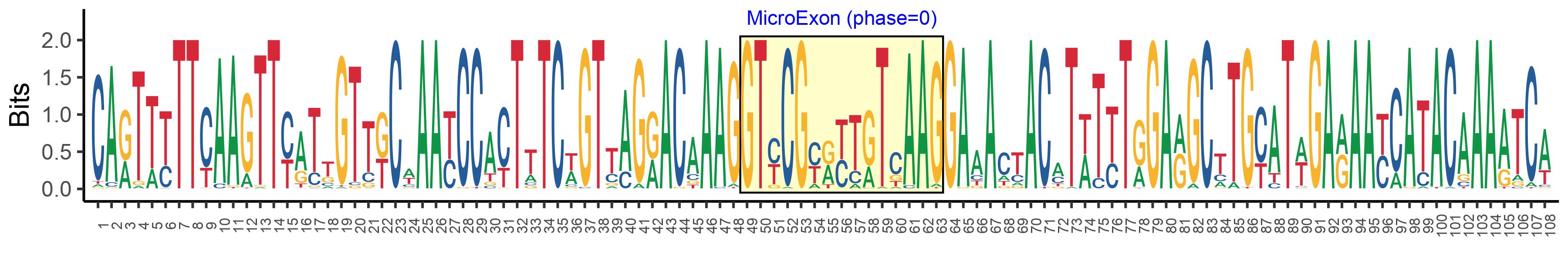 |
| Alignment of exons |  |
| Microexon DNA seq | GTCCGTGTAGTCAAG |
| Microexon Amino Acid seq | VRVVK |
| Microexon-tag DNA Seq | CAGTTTTTCAAGTTTATTGTTGCAAATCCGCTTTCGGTTAGGACTAAGGTCCGTGTAGTCAAGGAAACTACATATTTAGAGGCATGCATTGAGAATCACACAAAAACA |
| Microexon-tag Amino Acid Seq | QFFKFIVANPLSVRTKVRVVKETTYLEACIENHTKT |
| Microexon-tag spanning region | 3240727-3241664 |
| Microexon-tag prediction score | 0.9681 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | Potri.004G040500.1.v4.1x |
| Reference Transcript ID | Potri.004G040500.1.v4.1 |
| Gene ID | Potri.004G040500.v4.1 |
| Gene Name | NA |
| Transcript ID | Potri.004G040500.1.v4.1 |
| Protein ID | |
| Gene ID | Potri.004G040500.v4.1 |
| Gene Name | |
| Pfam domain motif | TRAPPC13_M |
| Motif E-value | 1.20E-20 |
| Motif start | 197 |
| Motif end | 313 |
| Protein seq | >Potri.004G040500.1.v4.1 MSTAPATQSLAFRVMRLCRPSFHVDTPLLLDPSDLILGEDIFDDPLAATHLPPLIDTHLTNPIDSSDLSYRSRFLLQNPS DSFGLSGLLVLPQSFGAIYLGETFCSYVSINNSSNFEVRDIVIKAEMQTERQRILLLDTSKTPVESIRASGRYDFIVEHD VKELGAHTLVCTALYTDGDGERKYLPQFFKFIVANPLSVRTKVRVVKETTYLEACIENHTKTNLYMDQVEFEPAPNWSAK ILKADEHKSKDNSPSREIFKPPVLVKSGGGIRNYLYQLSLSSHGSAESNVLGKLQITWRTNLGEPGRLQTQQILGTPITP KEIELHVAEVPSAINLDRPFLVHLNLTNQTDRELGPFEVWLSQDDTLDEKTVMINGLQTMELSQLEAFGSTDFYLNLIAT KLGVQKITGITVFDKSEKKTYAPLPDLEIFVDMH* |
| CDS seq | >Potri.004G040500.1.v4.1 ATGAGCACGGCACCAGCTACCCAATCACTAGCCTTCCGTGTAATGCGACTATGCCGTCCATCTTTCCACGTCGACACTCC TCTCCTTCTGGACCCCTCCGATCTCATTCTTGGTGAAGACATCTTCGACGATCCTCTCGCCGCCACTCACCTCCCTCCAC TCATCGACACCCACCTCACCAATCCGATCGACTCCTCCGATCTAAGTTACCGCTCTCGATTCCTCCTCCAAAACCCCTCC GATTCCTTCGGTCTCTCTGGCCTCCTCGTCCTCCCTCAATCCTTCGGGGCGATTTATTTGGGAGAGACGTTTTGCAGTTA CGTTAGCATTAATAACAGTTCTAATTTTGAAGTTAGAGATATTGTTATTAAGGCTGAAATGCAAACGGAGAGACAAAGGA TTTTACTGCTAGATACCTCAAAAACACCTGTTGAATCAATTCGAGCTAGTGGCCGTTACGATTTTATTGTTGAACATGAT GTGAAAGAACTCGGAGCTCATACGTTGGTTTGTACTGCTTTGTATACTGATGGTGATGGTGAAAGGAAATATCTGCCGCA GTTTTTCAAGTTTATTGTTGCAAATCCGCTTTCGGTTAGGACTAAGGTCCGTGTAGTCAAGGAAACTACATATTTAGAGG CATGCATTGAGAATCACACAAAAACAAACCTTTACATGGACCAAGTTGAGTTTGAGCCAGCTCCAAATTGGAGTGCAAAA ATTCTAAAAGCTGATGAACACAAATCTAAGGATAATTCTCCATCCAGGGAAATATTCAAGCCACCTGTCCTTGTTAAATC TGGTGGAGGAATTCGCAACTATCTTTACCAACTGTCATTGTCCTCACATGGTTCAGCAGAGAGTAATGTTCTTGGTAAGC TTCAGATAACATGGCGCACAAATTTAGGTGAACCTGGTCGCCTACAGACACAGCAAATACTTGGCACTCCCATCACGCCT AAGGAGATAGAGTTACATGTTGCTGAGGTTCCATCTGCCATCAACTTGGATAGACCATTTTTGGTACACTTGAATCTCAC AAACCAGACGGATAGAGAATTGGGCCCTTTTGAAGTTTGGTTGTCACAAGATGATACTCTCGATGAGAAAACTGTTATGA TTAATGGTCTCCAAACAATGGAATTATCCCAGTTGGAAGCATTTGGTAGCACAGATTTTTACTTGAACCTCATTGCTACT AAGCTTGGAGTCCAGAAAATAACAGGCATTACAGTATTTGACAAATCAGAGAAGAAAACTTATGCCCCATTGCCTGATTT AGAGATATTCGTGGACATGCATTGA |
Pt_4:3240958-3240972:+ does not have available information here.