
| Microexon ID | Gm_7:36371776-36371790:+ |
| Species | Glycine max | Coordinates | 7:36371776..36371790 |
| Microexon Cluster ID | MEP41 |
| Size | 15 |
| Phase | 0 |
| Pfam Domain Motif | DUF974 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,15,45 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | CARTTYTTCAAGTTYATTGTTKCWAAYCCACTTTCWGTTAGRACAAAGGTYCGYRYTRTCAAGGAAACTACMTWTYTRGARGCTTGYATWGARAAYCATACAAAATCA |
| Logo of Microexon-tag DNA Seq | 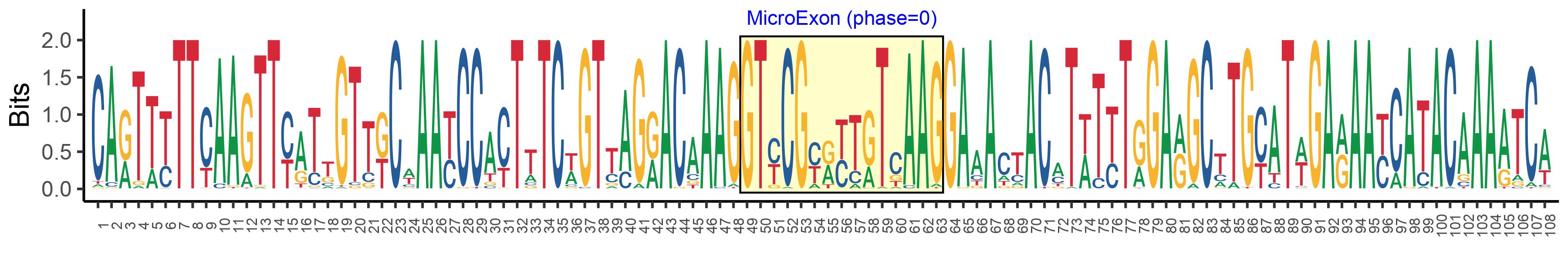 |
| Alignment of exons | 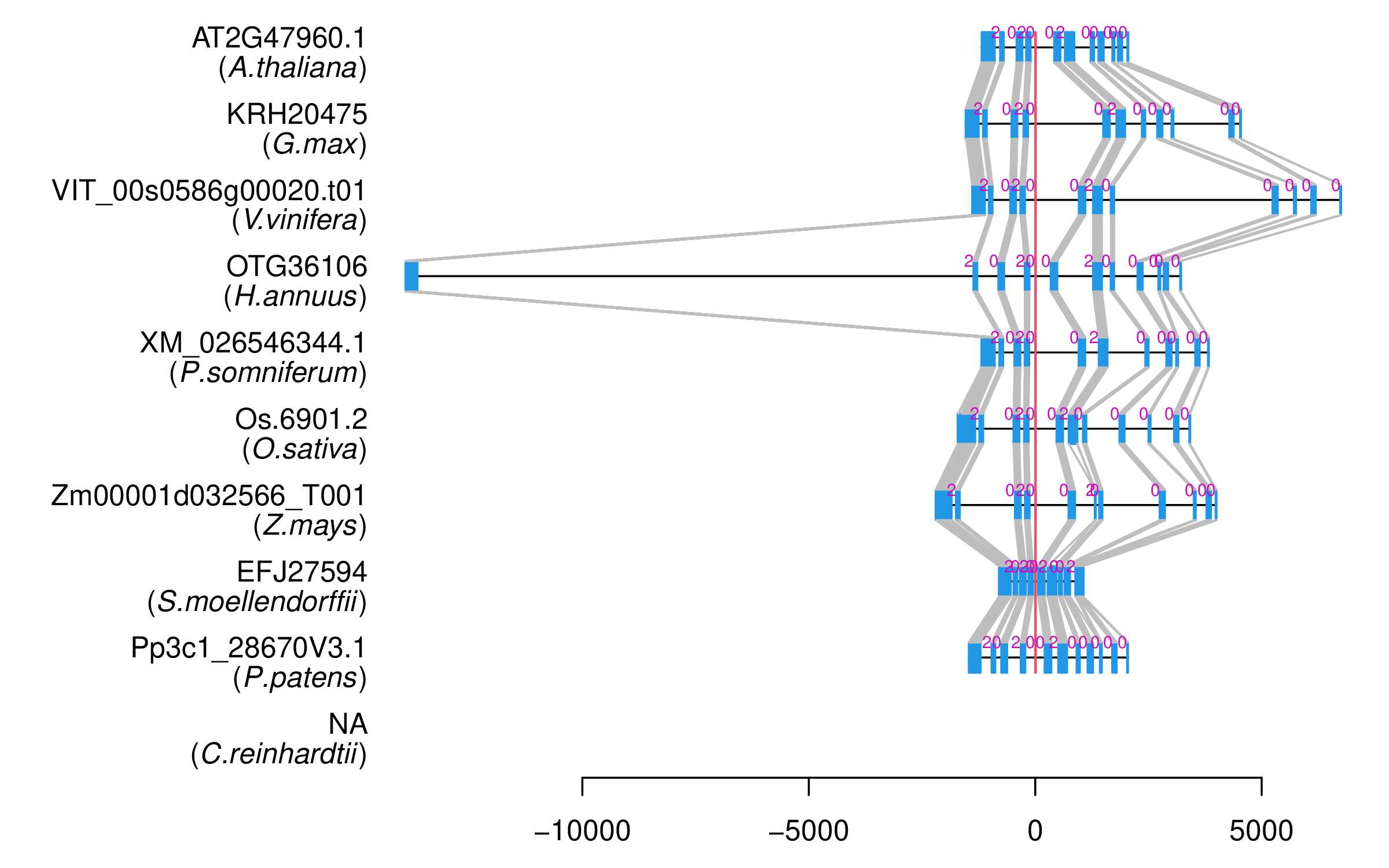 |
| Microexon DNA seq | GTCCGTGTTATCAAG |
| Microexon Amino Acid seq | VRVIK |
| Microexon-tag DNA Seq | CAATTTTTCAAGTTCATCGTTGCTAATCCACTTTCTGTTAGGACAAAGGTCCGTGTTATCAAGGAGACTACCTTTTTGGAAGCTTGTATTGAAAACCATACAAAATCA |
| Microexon-tag Amino Acid Seq | QFFKFIVANPLSVRTKVRVIKETTFLEACIENHTKS |
| Microexon-tag spanning region | 36371582-36373850 |
| Microexon-tag prediction score | 0.9826 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRH50013x |
| Reference Transcript ID | KRH50013 |
| Gene ID | GLYMA_07G195200 |
| Gene Name | NA |
| Transcript ID | KRH50013 |
| Protein ID | KRH50013 |
| Gene ID | GLYMA_07G195200 |
| Gene Name | NA |
| Pfam domain motif | DUF974 |
| Motif E-value | 1.9e-63 |
| Motif start | 85 |
| Motif end | 314 |
| Protein seq | >KRH50013 MSQGGQGGGGSHSLAFRVMRLCRPSFNVEPPLRLDPADLFAGEDLFDDPAANPPSFSSSDDSDSNYRNRFLLRHFSDAMG LSGLLVLPQSFGAIYLGETFCSYISINNSSNFEVRDVIIKAEIQTERLRILLLDTSKSPVETIRAGGRYDFIVEHDVKEL GPHTLVCTALYNDGDGERKYLPQFFKFIVANPLSVRTKVRVIKETTFLEACIENHTKSNLFMDQVDFEPAQYYSASILKG DGHHSEKDSPTRETFKPPILIRSGGGIYNYLYQLKTSSDGLPQTKVEGSNVLGKLQITWRTNLGEPGRLQTQQILGTTAT KKEIELQVVEVPSIINLQNPFMLKLNLTNQTDRELGPFEVSLSQNVSYGERAVMINGLQSMVLSEVQALGSTNFHLNLIA TKPGIQRITGITVFDTREMKSYEPLPDLEIFVDMD* |
| CDS seq | >KRH50013 ATGAGTCAGGGTGGTCAGGGAGGAGGAGGATCCCATTCCCTGGCGTTTCGCGTCATGCGTCTGTGCCGCCCTTCCTTCAA CGTCGAACCTCCCCTCCGCCTCGACCCCGCCGACCTCTTCGCCGGCGAGGACCTCTTCGACGACCCCGCCGCCAATCCCC CCTCTTTCTCCTCCTCCGACGACTCCGATTCCAACTACCGCAACCGCTTCCTCCTCCGCCACTTCTCCGACGCCATGGGC CTCTCCGGCCTCCTCGTTCTCCCTCAGTCTTTCGGAGCCATTTATTTGGGGGAGACTTTCTGCAGCTACATCAGCATAAA CAACAGCTCCAATTTTGAAGTCAGAGATGTCATTATCAAGGCTGAAATTCAAACAGAGAGACTGAGAATACTTCTTTTAG ACACATCAAAATCGCCAGTTGAAACGATACGTGCTGGTGGCCGTTATGATTTTATTGTAGAACATGATGTGAAAGAGCTT GGACCTCACACGCTGGTCTGCACTGCATTGTATAATGATGGTGACGGTGAGCGTAAATATCTTCCCCAATTTTTCAAGTT CATCGTTGCTAATCCACTTTCTGTTAGGACAAAGGTCCGTGTTATCAAGGAGACTACCTTTTTGGAAGCTTGTATTGAAA ACCATACAAAATCAAACCTTTTCATGGACCAAGTTGATTTTGAACCTGCTCAGTATTATAGTGCATCAATACTTAAAGGT GATGGGCACCATTCTGAGAAAGATAGTCCTACAAGGGAGACATTTAAGCCACCAATACTAATTAGATCTGGTGGAGGAAT TTACAACTATCTCTATCAATTGAAAACATCATCAGATGGTTTGCCTCAAACAAAAGTTGAAGGAAGTAATGTTCTTGGTA AACTCCAGATAACATGGCGAACAAATTTGGGTGAACCCGGTCGCCTGCAGACCCAGCAAATATTGGGGACAACAGCAACA AAGAAGGAGATTGAGTTGCAAGTTGTAGAGGTTCCATCTATAATTAACCTTCAAAATCCGTTCATGCTAAAATTGAATCT TACAAACCAGACAGATAGAGAGCTGGGTCCGTTTGAAGTTAGCTTATCTCAAAATGTTTCATATGGGGAGAGAGCTGTTA TGATTAATGGCCTTCAATCAATGGTTTTATCAGAGGTTCAGGCTTTAGGATCTACAAATTTCCACCTGAATCTCATAGCT ACTAAACCTGGAATTCAGAGAATTACGGGAATTACAGTTTTTGATACTAGGGAGATGAAATCTTATGAACCACTTCCAGA TTTGGAGATTTTTGTGGACATGGATTAA |
| Microexon DNA seq | GTCCGTGTTATCAAG |
| Microexon Amino Acid seq | VRVIK |
| Microexon-tag DNA Seq | CAATTTTTCAAGTTCATCGTTGCTAATCCACTTTCTGTTAGGACAAAGGTCCGTGTTATCAAGGAGACTACCTTTTTGGAAGCTTGTATTGAAAACCATACAAAATCA |
| Microexon-tag Amino Acid seq | QFFKFIVANPLSVRTKVRVIKETTFLEACIENHTKS |
| Transcript ID | KRH50013 |
| Gene ID | Gm.47361 |
| Gene Name | NA |
| Pfam domain motif | DUF974 |
| Motif E-value | 1.9e-63 |
| Motif start | 85 |
| Motif end | 314 |
| Protein seq | >KRH50013 MSQGGQGGGGSHSLAFRVMRLCRPSFNVEPPLRLDPADLFAGEDLFDDPAANPPSFSSSDDSDSNYRNRFLLRHFSDAMG LSGLLVLPQSFGAIYLGETFCSYISINNSSNFEVRDVIIKAEIQTERLRILLLDTSKSPVETIRAGGRYDFIVEHDVKEL GPHTLVCTALYNDGDGERKYLPQFFKFIVANPLSVRTKVRVIKETTFLEACIENHTKSNLFMDQVDFEPAQYYSASILKG DGHHSEKDSPTRETFKPPILIRSGGGIYNYLYQLKTSSDGLPQTKVEGSNVLGKLQITWRTNLGEPGRLQTQQILGTTAT KKEIELQVVEVPSIINLQNPFMLKLNLTNQTDRELGPFEVSLSQNVSYGERAVMINGLQSMVLSEVQALGSTNFHLNLIA TKPGIQRITGITVFDTREMKSYEPLPDLEIFVDMD* |
| CDS seq | >KRH50013 ATGAGTCAGGGTGGTCAGGGAGGAGGAGGATCCCATTCCCTGGCGTTTCGCGTCATGCGTCTGTGCCGCCCTTCCTTCAA CGTCGAACCTCCCCTCCGCCTCGACCCCGCCGACCTCTTCGCCGGCGAGGACCTCTTCGACGACCCCGCCGCCAATCCCC CCTCTTTCTCCTCCTCCGACGACTCCGATTCCAACTACCGCAACCGCTTCCTCCTCCGCCACTTCTCCGACGCCATGGGC CTCTCCGGCCTCCTCGTTCTCCCTCAGTCTTTCGGAGCCATTTATTTGGGGGAGACTTTCTGCAGCTACATCAGCATAAA CAACAGCTCCAATTTTGAAGTCAGAGATGTCATTATCAAGGCTGAAATTCAAACAGAGAGACTGAGAATACTTCTTTTAG ACACATCAAAATCGCCAGTTGAAACGATACGTGCTGGTGGCCGTTATGATTTTATTGTAGAACATGATGTGAAAGAGCTT GGACCTCACACGCTGGTCTGCACTGCATTGTATAATGATGGTGACGGTGAGCGTAAATATCTTCCCCAATTTTTCAAGTT CATCGTTGCTAATCCACTTTCTGTTAGGACAAAGGTCCGTGTTATCAAGGAGACTACCTTTTTGGAAGCTTGTATTGAAA ACCATACAAAATCAAACCTTTTCATGGACCAAGTTGATTTTGAACCTGCTCAGTATTATAGTGCATCAATACTTAAAGGT GATGGGCACCATTCTGAGAAAGATAGTCCTACAAGGGAGACATTTAAGCCACCAATACTAATTAGATCTGGTGGAGGAAT TTACAACTATCTCTATCAATTGAAAACATCATCAGATGGTTTGCCTCAAACAAAAGTTGAAGGAAGTAATGTTCTTGGTA AACTCCAGATAACATGGCGAACAAATTTGGGTGAACCCGGTCGCCTGCAGACCCAGCAAATATTGGGGACAACAGCAACA AAGAAGGAGATTGAGTTGCAAGTTGTAGAGGTTCCATCTATAATTAACCTTCAAAATCCGTTCATGCTAAAATTGAATCT TACAAACCAGACAGATAGAGAGCTGGGTCCGTTTGAAGTTAGCTTATCTCAAAATGTTTCATATGGGGAGAGAGCTGTTA TGATTAATGGCCTTCAATCAATGGTTTTATCAGAGGTTCAGGCTTTAGGATCTACAAATTTCCACCTGAATCTCATAGCT ACTAAACCTGGAATTCAGAGAATTACGGGAATTACAGTTTTTGATACTAGGGAGATGAAATCTTATGAACCACTTCCAGA TTTGGAGATTTTTGTGGACATGGATTAA |