
| Microexon ID | Zm_1:4418654-4418667:+ |
| Species | Zea mays | Coordinates | 1:4418654..4418667 |
| Microexon Cluster ID | MEP40 |
| Size | 14 |
| Phase | 1 |
| Pfam Domain Motif | SBP_bac_10 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 46,14,48 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GTYATCCCMYTRKCMAACTWYTCHRTBGAYRCCRMTTATTTTCCAGTKTCMTTCTTYGAGCTTYTAGGWYTRCTRGVRARCWTGAARGGCATMACATCAGAMWMRGTR |
| Logo of Microexon-tag DNA Seq | 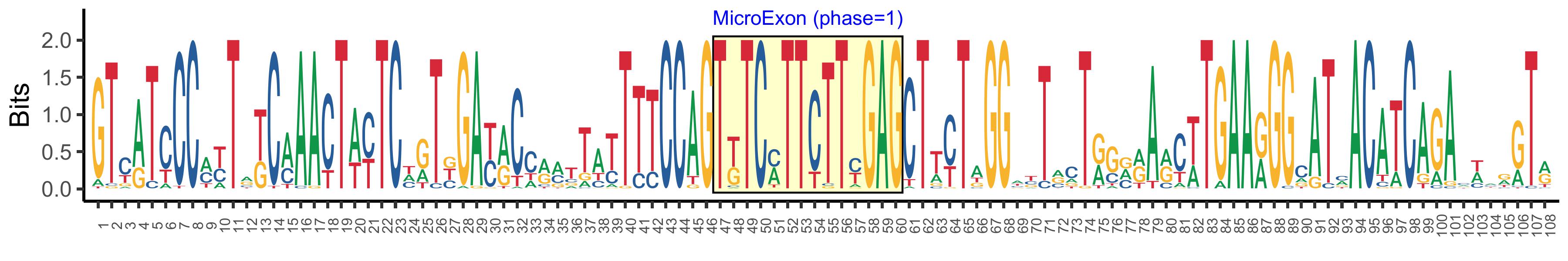 |
| Alignment of exons | 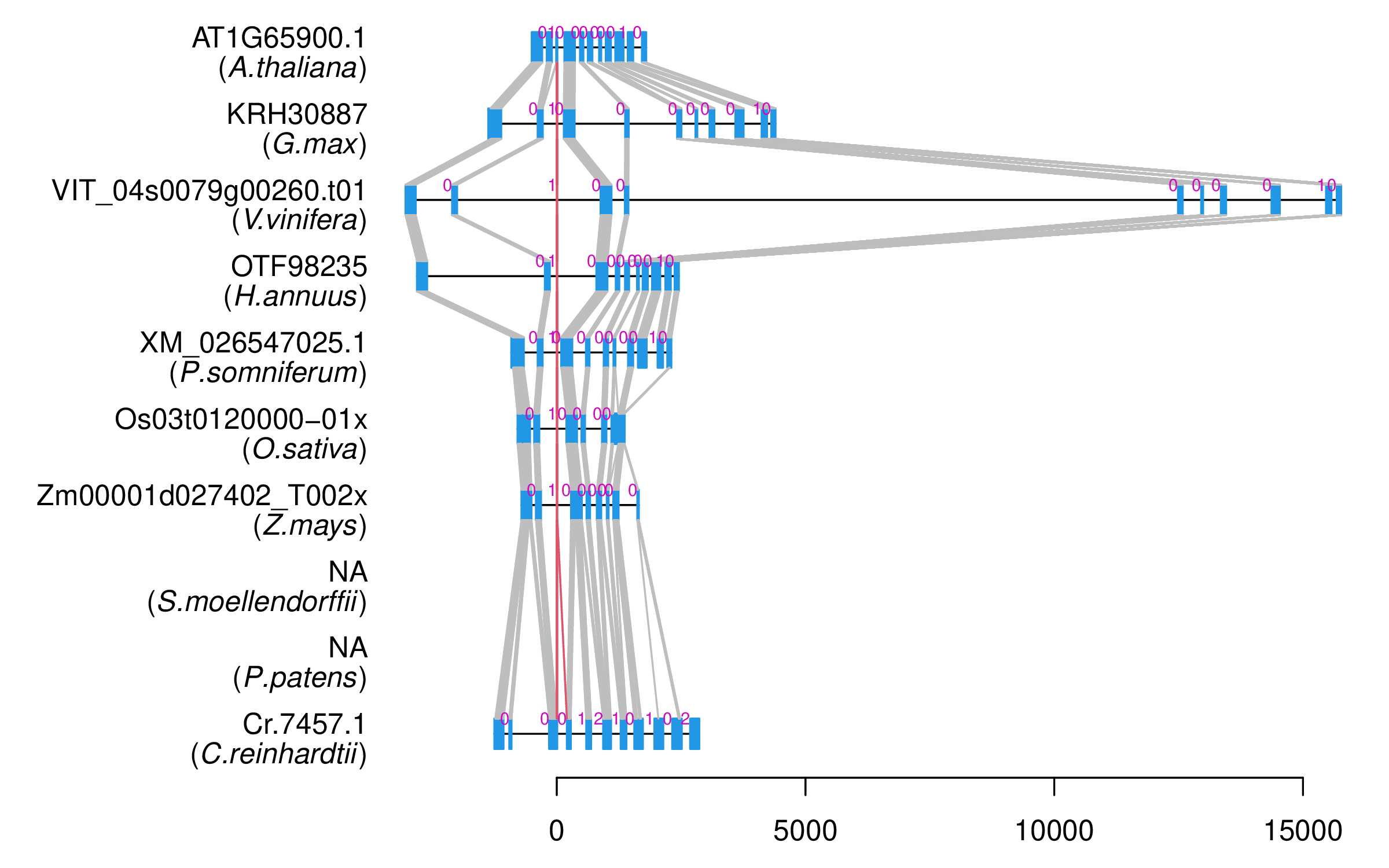 |
| Microexon DNA seq | TGTCATTCTTTGAG |
| Microexon Amino Acid seq | VSFFE |
| Microexon-tag DNA Seq | GTCATCCCCCTCGCCAGCTTCTCCGTCGACACCACGACCTCTCCAGTGTCATTCTTTGAGCTGCTTGGAGTGCTAGAGAGCTTGAAAGGAATAACATCGAACCAAGTC |
| Microexon-tag Amino Acid Seq | VIPLASFSVDTTTSPVSFFELLGVLESLKGITSNQV |
| Microexon-tag spanning region | 4418292-4418987 |
| Microexon-tag prediction score | 0.9395 |
| Overlapped with the annotated transcript (%) | 40 |
| New Transcript ID | Zm00001d027402_T002x |
| Reference Transcript ID | Zm00001d027402_T002 |
| Gene ID | Zm00001d027402 |
| Gene Name | NA |
Zm_1:4418654-4418667:+ does not have available information here.
| Microexon DNA seq | TGTCATTCTTTGAG |
| Microexon Amino Acid seq | VSFFE |
| Microexon-tag DNA Seq | GTCATCCCCCTCGCCAGCTTCTCCGTCGACACCACGACCTCTCCAGTGTCATTCTTTGAGCTGCTTGGAGTGCTAGAGAGCTTGAAAGGAATAACATCGAACCAAGTC |
| Microexon-tag Amino Acid seq | VIPLASFSVDTTTSPVSFFELLGVLESLKGITSNQV |
| Transcript ID | Zm.135.1 |
| Gene ID | Zm.135 |
| Gene Name | NA |
| Pfam domain motif | Unknown |
| Motif E-value | NA |
| Motif start | NA |
| Motif end | NA |
| Protein seq | >Zm.135.1 MAAVRRCCLLAAVLLWSALASAATAAPAPVVAGPVSRVEDATMFRIYYGQSFKVIKNSGDGKSYLLMQNTSKMASKTKYC TGRIKSFVIPLASFSVDTTTSPVSFFELLGVLESLKGITSNQVASQCVLQSYTSGNAQLVNRTDVQTLSQFTAHFISDTD EDRGCNFAAYVPLEEGTPLQMAEWIKYLGTFTNSENRANAVYDAVKTNYLCLSKVAANLGTRFKPVVAWIEFTQGMWTFV KESYILQYVTDAGAEIVDATITDKRFNSSDPEDMDNFHAILCTVDVVIDQTYALEPAEYKLSTFLENINVSHDSCYSFVS NRSIWRFDKRIGASGTLDWSDGAISQPQLVLGDLIEIFFPTGNYITIYFRNLAKDEGVTEIGPEMCTRSISTPMDPIILS CE* |
| CDS seq | >Zm.135.1 ATGGCCGCTGTTCGTAGATGCTGCCTGCTGGCCGCGGTGCTGCTGTGGTCGGCGCTCGCTTCAGCGGCCACGGCCGCGCC GGCGCCTGTGGTGGCCGGGCCGGTGTCCAGGGTGGAGGACGCCACCATGTTCCGGATATACTACGGTCAGAGCTTCAAGG TCATCAAGAACTCCGGCGATGGCAAGAGCTACCTTCTCATGCAGAACACGTCCAAGATGGCATCCAAGACGAAGTACTGC ACGGGGAGGATTAAGTCGTTCGTCATCCCCCTCGCCAGCTTCTCCGTCGACACCACGACCTCTCCAGTGTCATTCTTTGA GCTGCTTGGAGTGCTAGAGAGCTTGAAAGGAATAACATCGAACCAAGTCGCTTCTCAGTGCGTGCTCCAGTCATATACAA GTGGAAATGCCCAGCTTGTGAATAGAACTGATGTGCAGACACTCTCGCAGTTTACTGCGCATTTCATAAGCGATACAGAT GAAGACAGAGGTTGCAACTTTGCAGCATATGTACCCTTAGAAGAGGGCACACCATTGCAGATGGCTGAGTGGATCAAGTA CCTGGGAACTTTCACAAACTCTGAAAACAGGGCTAATGCTGTCTACGATGCAGTAAAGACAAACTACCTGTGCTTAAGCA AAGTAGCTGCAAACCTTGGCACAAGATTCAAGCCTGTAGTAGCGTGGATTGAATTCACACAGGGAATGTGGACATTTGTT AAAGAAAGTTACATCTTACAGTATGTCACAGATGCAGGAGCAGAAATTGTTGATGCAACTATTACGGATAAGAGATTTAA TAGTTCAGATCCAGAGGATATGGATAACTTCCATGCTATTCTGTGTACAGTGGACGTGGTCATAGATCAGACATATGCTT TGGAGCCTGCAGAGTACAAACTATCCACCTTTTTGGAGAATATTAATGTTAGCCACGACTCCTGTTACAGTTTTGTATCA AATCGCAGCATATGGAGATTTGACAAAAGGATAGGAGCTTCTGGGACACTTGATTGGTCCGATGGAGCTATCTCACAGCC TCAGTTGGTTCTTGGAGATCTCATAGAAATTTTCTTCCCAACGGGGAACTACATAACAATTTACTTCAGGAATCTTGCAA AGGATGAAGGGGTCACAGAGATTGGTCCTGAAATGTGCACTAGGAGCATATCCACACCCATGGATCCTATAATCCTGTCA TGTGAATGA |