
| Microexon ID | Gm_18:3757877-3757890:- |
| Species | Glycine max | Coordinates | 18:3757877..3757890 |
| Microexon Cluster ID | MEP40 |
| Size | 14 |
| Phase | 1 |
| Pfam Domain Motif | SBP_bac_10 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 46,14,48 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GTYATCCCMYTRKCMAACTWYTCHRTBGAYRCCRMTTATTTTCCAGTKTCMTTCTTYGAGCTTYTAGGWYTRCTRGVRARCWTGAARGGCATMACATCAGAMWMRGTR |
| Logo of Microexon-tag DNA Seq | 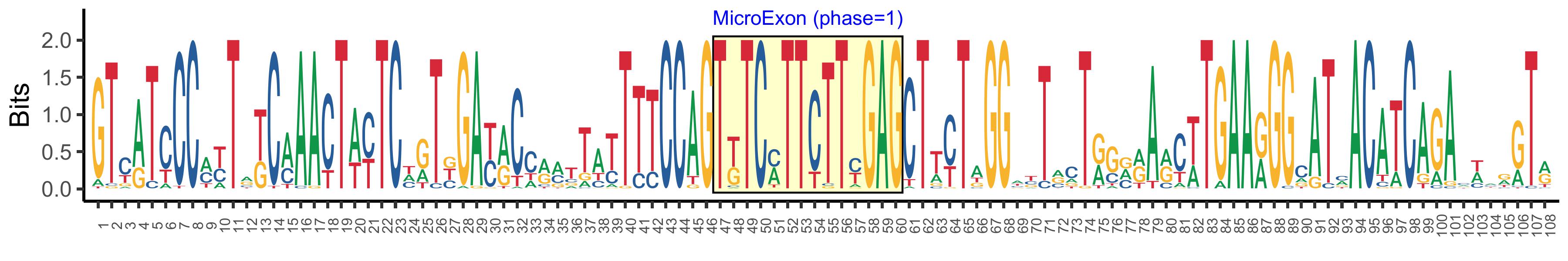 |
| Alignment of exons | 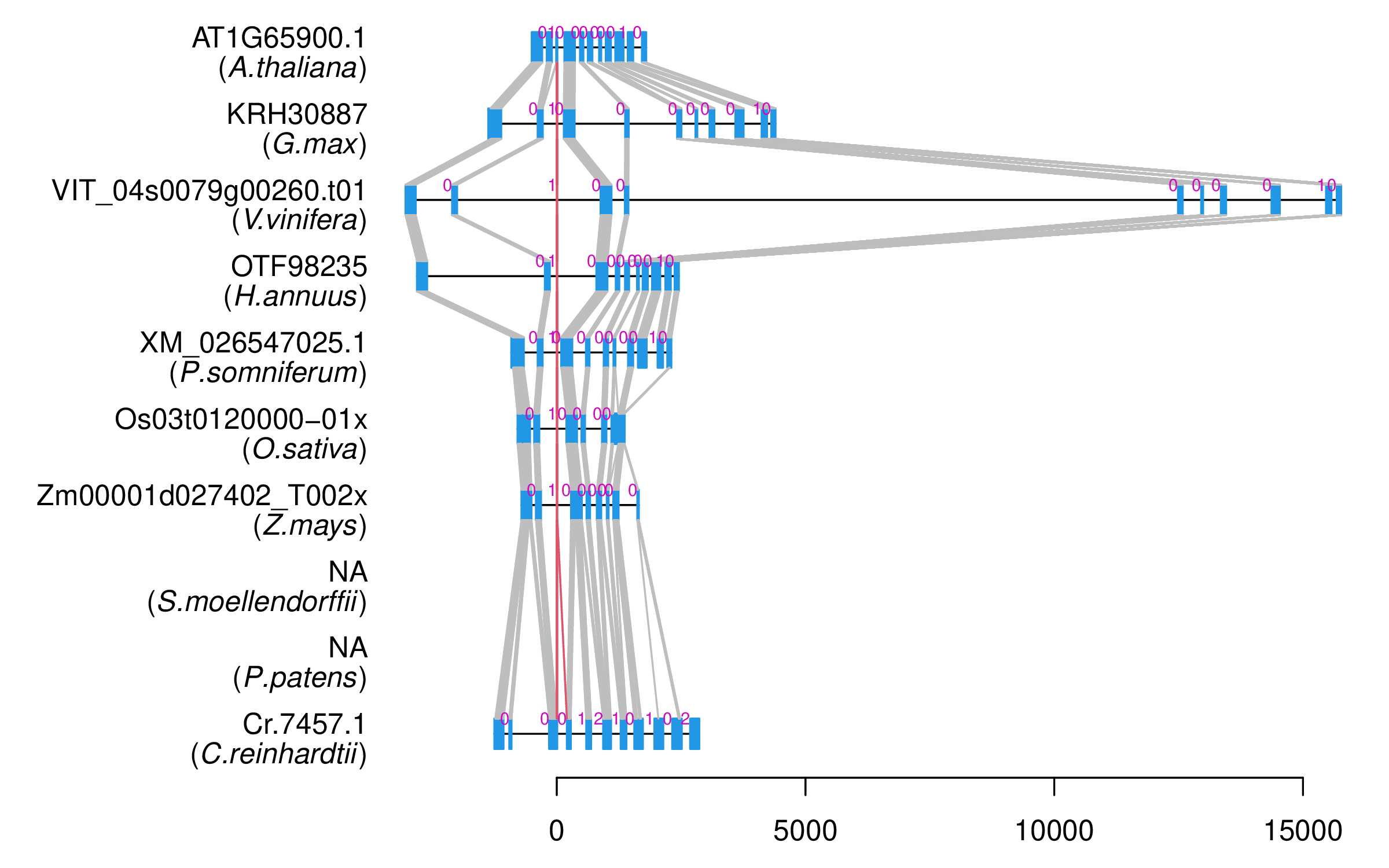 |
| Microexon DNA seq | TCTCCTTCTTAGAG |
| Microexon Amino Acid seq | VSFLE |
| Microexon-tag DNA Seq | GTCATACCATTGTCAAACTATTCTGTCGATACAACTTATTTTCCGGTCTCCTTCTTAGAGCTTTTAGGTTTAGTGGAGAGCTTGAAAGGCATAACCTCAGACTATGTG |
| Microexon-tag Amino Acid Seq | VIPLSNYSVDTTYFPVSFLELLGLVESLKGITSDYV |
| Microexon-tag spanning region | 3757703-3758179 |
| Microexon-tag prediction score | 0.9502 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRG98001x |
| Reference Transcript ID | KRG98001 |
| Gene ID | GLYMA_18G043800 |
| Gene Name | NA |
| Transcript ID | KRG98001 |
| Protein ID | KRG98001 |
| Gene ID | GLYMA_18G043800 |
| Gene Name | NA |
| Pfam domain motif | Unknown |
| Motif E-value | NA |
| Motif start | NA |
| Motif end | NA |
| Protein seq | >KRG98001 MDSAPSCSWLQAFSFLALVWLFNCGCVHGATPAVKVGNFSKVEDAGNFHIYYGQTFKVIKNSADGQSYLLLQNNSRIASR TKYCTSRIKSFVIPLSNYSVDTTYFPVSFLELLGLVESLKGITSDYVASPCVLKLYEGGQIELFNNSDYQKLAEFSSYFL SDTDQQPACNFATFVPFIEDIPLQRAEWIKFMGAFANVEARANQVYTAVKANYLCLAKIATSRTTFKPTVAWMRYKNGLW SFTQEKYQLKYVQDAGGEILGANKNTYNVSDPDDLEEFHAILCTVEVVIDETLTSDPVNYTFSTFIQNLNVEDRSCFSFI SNTSLWRYDKRVYNSVALDWYNGAVSQPQLALADLIEVLFPTENYTTTYFRNIAKAEVPINIGLEMCDRDTSTAMEPTIV ACG* |
| CDS seq | >KRG98001 ATGGATTCAGCTCCTTCTTGTTCTTGGCTACAAGCCTTCTCCTTCTTAGCATTGGTTTGGTTATTTAACTGTGGATGTGT CCATGGAGCCACCCCAGCAGTGAAAGTAGGTAACTTTTCAAAGGTGGAAGATGCTGGAAACTTCCATATTTACTATGGCC AGACCTTCAAAGTCATTAAGAATTCTGCTGATGGCCAGAGCTACCTTCTTCTCCAGAATAATTCAAGGATTGCATCAAGG ACTAAATATTGCACATCAAGAATCAAGTCATTTGTCATACCATTGTCAAACTATTCTGTCGATACAACTTATTTTCCGGT CTCCTTCTTAGAGCTTTTAGGTTTAGTGGAGAGCTTGAAAGGCATAACCTCAGACTATGTGGCTTCTCCATGTGTGTTGA AACTGTACGAAGGAGGACAGATAGAATTGTTCAATAATAGTGATTACCAAAAGCTTGCAGAGTTCTCTTCATACTTTCTT AGTGACACTGATCAGCAGCCAGCTTGCAATTTTGCAACTTTTGTTCCATTTATAGAGGATATCCCTTTGCAGAGAGCAGA ATGGATCAAATTCATGGGAGCTTTTGCAAATGTTGAAGCTAGAGCCAATCAAGTCTATACAGCAGTTAAGGCAAACTATT TGTGCTTGGCTAAAATTGCTACAAGTAGGACAACATTCAAGCCAACGGTAGCTTGGATGAGGTATAAAAATGGTCTTTGG TCTTTTACACAGGAAAAGTATCAATTGAAGTACGTGCAAGATGCAGGCGGAGAGATTTTGGGTGCCAACAAGAACACTTA CAATGTCTCTGATCCTGATGACTTGGAGGAATTTCATGCCATCCTATGTACCGTGGAAGTAGTCATTGATGAAACACTAA CATCTGATCCAGTCAACTACACCTTTTCAACATTTATCCAAAATCTAAACGTTGAAGATCGTTCTTGTTTTTCTTTTATT TCAAACACAAGTCTATGGAGATATGACAAAAGGGTTTATAATTCTGTAGCTCTTGACTGGTATAATGGAGCAGTGTCCCA ACCTCAATTGGCACTAGCAGATCTTATTGAAGTTTTATTTCCCACTGAAAATTACACAACGACCTATTTTAGGAACATTG CAAAGGCAGAAGTACCTATAAATATTGGTCTTGAAATGTGTGACAGGGACACATCCACAGCAATGGAGCCTACCATAGTA GCATGTGGATGA |
| Microexon DNA seq | TCTCCTTCTTAGAG |
| Microexon Amino Acid seq | VSFLE |
| Microexon-tag DNA Seq | GTCATACCATTGTCAAACTATTCTGTCGATACAACTTATTTTCCGGTCTCCTTCTTAGAGCTTTTAGGTTTAGTGGAGAGCTTGAAAGGCATAACCTCAGACTATGTG |
| Microexon-tag Amino Acid seq | VIPLSNYSVDTTYFPVSFLELLGLVESLKGITSDYV |
| Transcript ID | KRG98002 |
| Gene ID | Gm.24416 |
| Gene Name | NA |
| Pfam domain motif | Unknown |
| Motif E-value | NA |
| Motif start | NA |
| Motif end | NA |
| Protein seq | >KRG98002 MDSAPSCSWLQAFSFLALVWLFNCGCVHGATPAVKVGNFSKVEDAGNFHIYYGQTFKVIKNSADGQSYLLLQNNSRIASR TKYCTSRIKSFVIPLSNYSVDTTYFPVSFLELLGLVESLKGITSDYVASPCVLKLYEGGQIELFNNSDYQKLAEFSSYFL SDTDQQPACNFATFVPFIEDIPLQRAEWIKFMGAFANVEARANQVYTAVKANYLCLAKIATSRTTFKPTVAWMRYKNGLW SFTQEKYQLKYVQDAGGEILGANKNTYNVSDPDDLEEFHAILCTVEVVIDETLTSDPVNYTFSTFIQNLNVEDRSCFSFI SNTSLWRYDKRVYNSVALDWYNGAVSQPQLALADLIEVLFPTENYTTTYFRNIAKAEVPINIGLEMCDRDTSTAMEPTIV ACG* |
| CDS seq | >KRG98002 ATGGATTCAGCTCCTTCTTGTTCTTGGCTACAAGCCTTCTCCTTCTTAGCATTGGTTTGGTTATTTAACTGTGGATGTGT CCATGGAGCCACCCCAGCAGTGAAAGTAGGTAACTTTTCAAAGGTGGAAGATGCTGGAAACTTCCATATTTACTATGGCC AGACCTTCAAAGTCATTAAGAATTCTGCTGATGGCCAGAGCTACCTTCTTCTCCAGAATAATTCAAGGATTGCATCAAGG ACTAAATATTGCACATCAAGAATCAAGTCATTTGTCATACCATTGTCAAACTATTCTGTCGATACAACTTATTTTCCGGT CTCCTTCTTAGAGCTTTTAGGTTTAGTGGAGAGCTTGAAAGGCATAACCTCAGACTATGTGGCTTCTCCATGTGTGTTGA AACTGTACGAAGGAGGACAGATAGAATTGTTCAATAATAGTGATTACCAAAAGCTTGCAGAGTTCTCTTCATACTTTCTT AGTGACACTGATCAGCAGCCAGCTTGCAATTTTGCAACTTTTGTTCCATTTATAGAGGATATCCCTTTGCAGAGAGCAGA ATGGATCAAATTCATGGGAGCTTTTGCAAATGTTGAAGCTAGAGCCAATCAAGTCTATACAGCAGTTAAGGCAAACTATT TGTGCTTGGCTAAAATTGCTACAAGTAGGACAACATTCAAGCCAACGGTAGCTTGGATGAGGTATAAAAATGGTCTTTGG TCTTTTACACAGGAAAAGTATCAATTGAAGTACGTGCAAGATGCAGGCGGAGAGATTTTGGGTGCCAACAAGAACACTTA CAATGTCTCTGATCCTGATGACTTGGAGGAATTTCATGCCATCCTATGTACCGTGGAAGTAGTCATTGATGAAACACTAA CATCTGATCCAGTCAACTACACCTTTTCAACATTTATCCAAAATCTAAACGTTGAAGATCGTTCTTGTTTTTCTTTTATT TCAAACACAAGTCTATGGAGATATGACAAAAGGGTTTATAATTCTGTAGCTCTTGACTGGTATAATGGAGCAGTGTCCCA ACCTCAATTGGCACTAGCAGATCTTATTGAAGTTTTATTTCCCACTGAAAATTACACAACGACCTATTTTAGGAACATTG CAAAGGCAGAAGTACCTATAAATATTGGTCTTGAAATGTGTGACAGGGACACATCCACAGCAATGGAGCCTACCATAGTA GCATGTGGATGA |