
| Microexon ID | Ta_5A:510279180-510279193:- |
| Species | Triticum Aestivum | Coordinates | 5A:510279180..510279193 |
| Microexon Cluster ID | MEP37 |
| Size | 14 |
| Phase | 1 |
| Pfam Domain Motif | Trigger_N |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 46,14,48 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GYTSSTRCWGCACMRCCARTTCCWGGHTTYCGMAGRSWRAAAGGAGGRAAAACAYCARAKRTACCMARARRCWTYCTWYTRSARATKMTTGGWSMAKMMMRAGTBAMS |
| Logo of Microexon-tag DNA Seq | 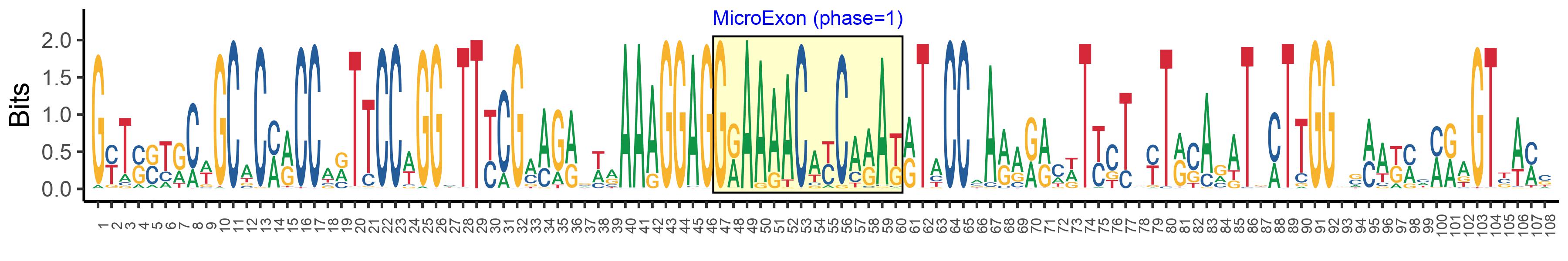 |
| Alignment of exons |  |
| Microexon DNA seq | GGAAAACTCCAAAT |
| Microexon Amino Acid seq | GKTPN |
| Microexon-tag DNA Seq | ATTGCCGCGGCACAACCACTTCCAGGATTTCGGCGATTGAAAGGAGGGAAAACTCCAAATGTACCAAAAGAAGTCGCTCTGCACTTGATTGGACCATCAAAAGTGAAG |
| Microexon-tag Amino Acid Seq | IAAAQPLPGFRRLKGGKTPNVPKEVALHLIGPSKVK |
| Microexon-tag spanning region | 510279027-510279630 |
| Microexon-tag prediction score | 0.9574 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | TraesCS5A02G301200.1x |
| Reference Transcript ID | TraesCS5A02G301200.1 |
| Gene ID | TraesCS5A02G301200 |
| Gene Name | NA |
| Transcript ID | TraesCS5A02G301200.1 |
| Protein ID | |
| Gene ID | TraesCS5A02G301200 |
| Gene Name | |
| Pfam domain motif | Trigger_N |
| Motif E-value | 1.50E-09 |
| Motif start | 13 |
| Motif end | 134 |
| Protein seq | >TraesCS5A02G301200.1 MQAPVAFKDFHVSVLTEEDGVIEIRVTVSSKMTVSVFEKVLSKHIAAAQPLPGFRRLKGGKTPNVPKEVALHLIGPSKVK KAAIKKIINRAVAEYVEKENLDASKNLKVLQSYEELEATFEPGKEFCFDAAVHLTGS* |
| CDS seq | >TraesCS5A02G301200.1 ATGCAAGCACCGGTAGCCTTTAAAGATTTTCATGTTTCTGTGCTTACCGAAGAAGATGGAGTGATAGAGATACGAGTAAC TGTTAGCAGTAAAATGACGGTTTCTGTCTTTGAGAAAGTCTTATCAAAGCATATTGCCGCGGCACAACCACTTCCAGGAT TTCGGCGATTGAAAGGAGGGAAAACTCCAAATGTACCAAAAGAAGTCGCTCTGCACTTGATTGGACCATCAAAAGTGAAG AAAGCAGCCATCAAGAAAATCATCAATCGCGCAGTCGCTGAATATGTTGAAAAGGAGAACCTTGACGCGTCGAAGAACCT GAAGGTTCTGCAAAGCTACGAGGAGCTCGAGGCCACGTTCGAACCTGGAAAAGAGTTCTGCTTTGACGCGGCTGTCCATC TAACAGGAAGCTGA |
Ta_5A:510279180-510279193:- does not have available information here.