
| Microexon ID | Vv_6:634354-634367:+ |
| Species | Vistis vinifera | Coordinates | 6:634354..634367 |
| Microexon Cluster ID | MEP36 |
| Size | 14 |
| Phase | 1 |
| Pfam Domain Motif | EFP_N |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 15,31,14,48 |
| Microexon location in the Microexon-tag | 3 |
| Microexon-tag DNA Seq | KTYYGCGGCTCYGATGTGARRCYWGGRAAYGTCATTGAAARAAAAGGAMRBATTTAYSAGGTKRTAAARGCASAACAYWCWMMTCAAGGAAGAGGAGGAGCYAYWATA |
| Logo of Microexon-tag DNA Seq | 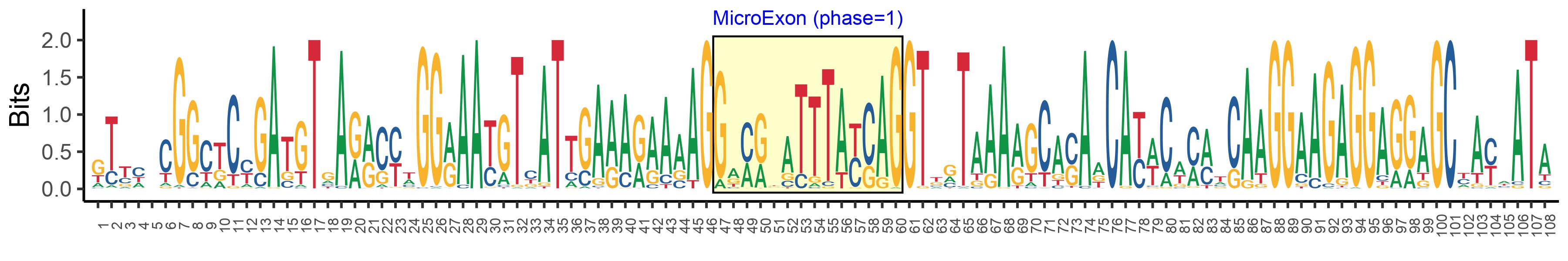 |
| Alignment of exons | 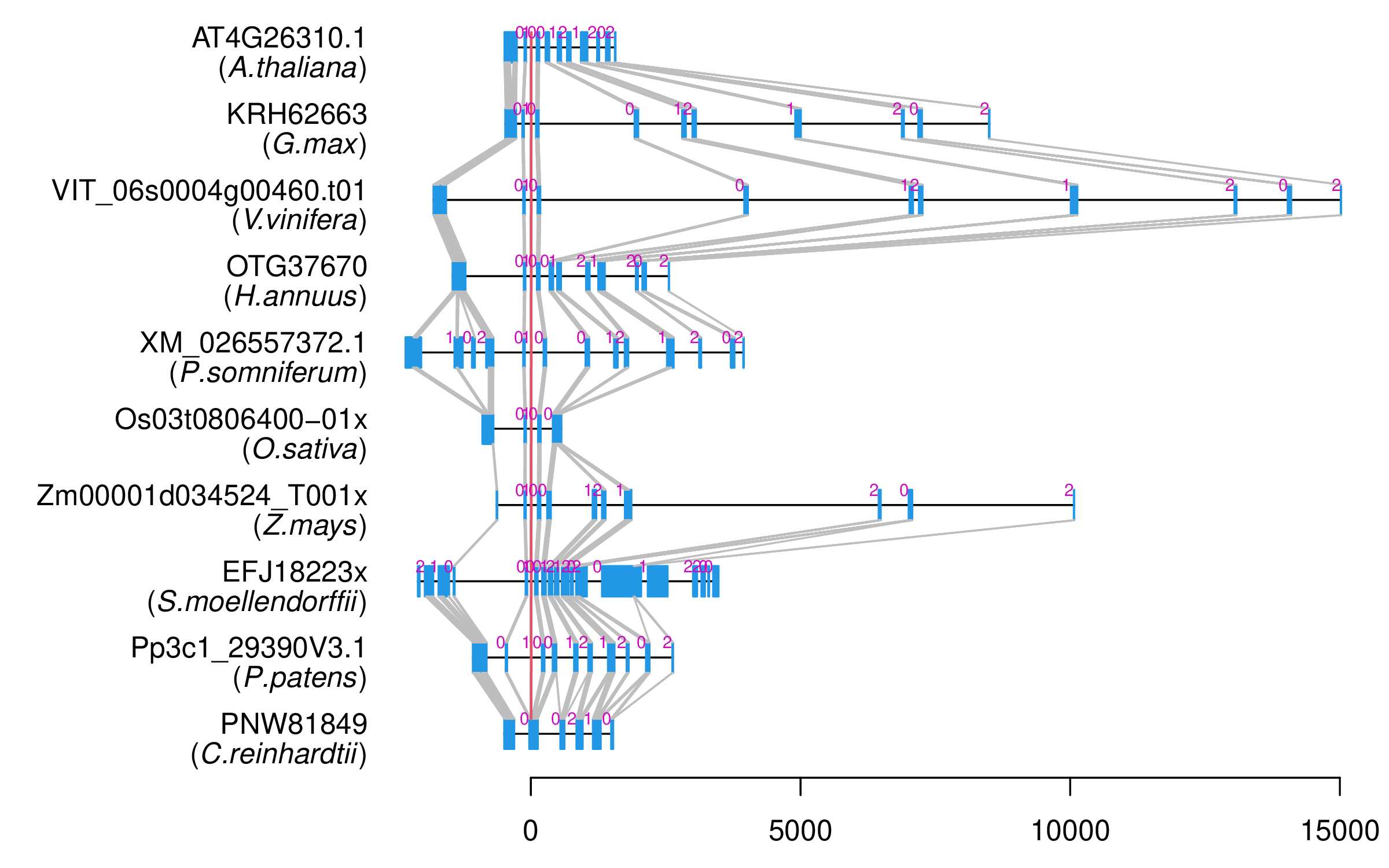 |
| Microexon DNA seq | GACGCATATACCAG |
| Microexon Amino Acid seq | GRIYQ |
| Microexon-tag DNA Seq | TTTCTCGGCTCCGATGTCAGACCTGGAAATTGCATCAGAAGAAAAGGACGCATATACCAGGTTTTAAAAGCACAACACACACAGCATGGAAGAGGAGGAGCAACAATA |
| Microexon-tag Amino Acid Seq | FLGSDVRPGNCIRRKGRIYQVLKAQHTQHGRGGATI |
| Microexon-tag spanning region | 632769-634525 |
| Microexon-tag prediction score | 0.9446 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | VIT_06s0004g00460.t01x |
| Reference Transcript ID | VIT_06s0004g00460.t01 |
| Gene ID | VIT_06s0004g00460 |
| Gene Name | NA |
| Transcript ID | VIT_06s0004g00460.t01 |
| Protein ID | VIT_06s0004g00460.t01 |
| Gene ID | VIT_06s0004g00460 |
| Gene Name | NA |
| Pfam domain motif | EFP_N |
| Motif E-value | 4.1e-17 |
| Motif start | 78 |
| Motif end | 132 |
| Protein seq | >VIT_06s0004g00460.t01 MRALLCRRLSRSLYETAASSSSFSSSFTSSSSSCFRRLGFRTLSSLTSSSNHTPLSRTLLASSPWSATQLRGAKFLGSDV RPGNCIRRKGRIYQVLKAQHTQHGRGGATIQVELRDIDSGSKVNEKFRTDETIESVFVEEKSFLYLYTDDDSIVLMEPGT FSQMEVSKDLFGKAAAYLKDDMKVTLHLCDGKPTSASVPKRVTCTVVEAQVPVKGSTATPQYKRVSLDNGLTVLAPPFVL AGDSIVIDTTDDSYITRA* |
| CDS seq | >VIT_06s0004g00460.t01 ATGCGAGCTCTGCTCTGCCGGCGACTCTCTCGATCTCTCTACGAAACCGCGGCTTCATCATCATCATTTTCTTCTTCTTT TACTTCTTCTTCATCATCATGTTTTCGTCGGCTCGGATTCAGAACTCTAAGCTCACTCACATCGTCGTCAAACCACACGC CTCTCTCTCGAACCCTACTCGCATCATCCCCGTGGTCCGCCACTCAACTGCGCGGCGCCAAGTTTCTCGGCTCCGATGTC AGACCTGGAAATTGCATCAGAAGAAAAGGACGCATATACCAGGTTTTAAAAGCACAACACACACAGCATGGAAGAGGAGG AGCAACAATACAGGTGGAGCTCCGTGATATTGATAGTGGAAGCAAAGTAAATGAAAAGTTTCGTACTGATGAGACTATTG AAAGTGTATTTGTTGAGGAAAAATCTTTCTTATATTTGTATACTGATGACGACAGCATAGTGCTGATGGAGCCTGGCACA TTTTCACAAATGGAAGTCTCAAAGGACTTGTTTGGCAAAGCTGCTGCATATCTAAAAGATGACATGAAAGTTACACTGCA TTTATGTGATGGAAAGCCAACATCTGCATCAGTTCCAAAACGCGTGACATGCACTGTTGTGGAAGCACAAGTGCCTGTGA AAGGGAGTACAGCCACACCTCAGTATAAAAGGGTTTCGCTGGACAATGGCCTTACAGTGTTGGCACCACCTTTTGTCCTT GCTGGTGACTCAATAGTAATTGATACAACTGATGACTCTTATATCACCAGGGCATAG |
| Microexon DNA seq | GACGCATATACCAG |
| Microexon Amino Acid seq | GRIYQ |
| Microexon-tag DNA Seq | TTTCTCGGCTCCGATGTCAGACCTGGAAATTGCATCAGAAGAAAAGGACGCATATACCAGGTTTTAAAAGCACAACACACACAGCATGGAAGAGGAGGAGCAACAATA |
| Microexon-tag Amino Acid seq | FLGSDVRPGNCIRRKGRIYQVLKAQHTQHGRGGATI |
| Transcript ID | VIT_06s0004g00460.t01 |
| Gene ID | Vv.26317 |
| Gene Name | NA |
| Pfam domain motif | EFP_N |
| Motif E-value | 4.1e-17 |
| Motif start | 78 |
| Motif end | 132 |
| Protein seq | >VIT_06s0004g00460.t01 MRALLCRRLSRSLYETAASSSSFSSSFTSSSSSCFRRLGFRTLSSLTSSSNHTPLSRTLLASSPWSATQLRGAKFLGSDV RPGNCIRRKGRIYQVLKAQHTQHGRGGATIQVELRDIDSGSKVNEKFRTDETIESVFVEEKSFLYLYTDDDSIVLMEPGT FSQMEVSKDLFGKAAAYLKDDMKVTLHLCDGKPTSASVPKRVTCTVVEAQVPVKGSTATPQYKRVSLDNGLTVLAPPFVL AGDSIVIDTTDDSYITRA* |
| CDS seq | >VIT_06s0004g00460.t01 ATGCGAGCTCTGCTCTGCCGGCGACTCTCTCGATCTCTCTACGAAACCGCGGCTTCATCATCATCATTTTCTTCTTCTTT TACTTCTTCTTCATCATCATGTTTTCGTCGGCTCGGATTCAGAACTCTAAGCTCACTCACATCGTCGTCAAACCACACGC CTCTCTCTCGAACCCTACTCGCATCATCCCCGTGGTCCGCCACTCAACTGCGCGGCGCCAAGTTTCTCGGCTCCGATGTC AGACCTGGAAATTGCATCAGAAGAAAAGGACGCATATACCAGGTTTTAAAAGCACAACACACACAGCATGGAAGAGGAGG AGCAACAATACAGGTGGAGCTCCGTGATATTGATAGTGGAAGCAAAGTAAATGAAAAGTTTCGTACTGATGAGACTATTG AAAGTGTATTTGTTGAGGAAAAATCTTTCTTATATTTGTATACTGATGACGACAGCATAGTGCTGATGGAGCCTGGCACA TTTTCACAAATGGAAGTCTCAAAGGACTTGTTTGGCAAAGCTGCTGCATATCTAAAAGATGACATGAAAGTTACACTGCA TTTATGTGATGGAAAGCCAACATCTGCATCAGTTCCAAAACGCGTGACATGCACTGTTGTGGAAGCACAAGTGCCTGTGA AAGGGAGTACAGCCACACCTCAGTATAAAAGGGTTTCGCTGGACAATGGCCTTACAGTGTTGGCACCACCTTTTGTCCTT GCTGGTGACTCAATAGTAATTGATACAACTGATGACTCTTATATCACCAGGGCATAG |