
| Microexon ID | Ps_NC_039368.1:39028757-39028770:+ |
| Species | Papaver somniferum | Coordinates | NC_039368.1:39028757..39028770 |
| Microexon Cluster ID | MEP36 |
| Size | 14 |
| Phase | 1 |
| Pfam Domain Motif | EFP_N |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 15,31,14,48 |
| Microexon location in the Microexon-tag | 3 |
| Microexon-tag DNA Seq | KTYYGCGGCTCYGATGTGARRCYWGGRAAYGTCATTGAAARAAAAGGAMRBATTTAYSAGGTKRTAAARGCASAACAYWCWMMTCAAGGAAGAGGAGGAGCYAYWATA |
| Logo of Microexon-tag DNA Seq | 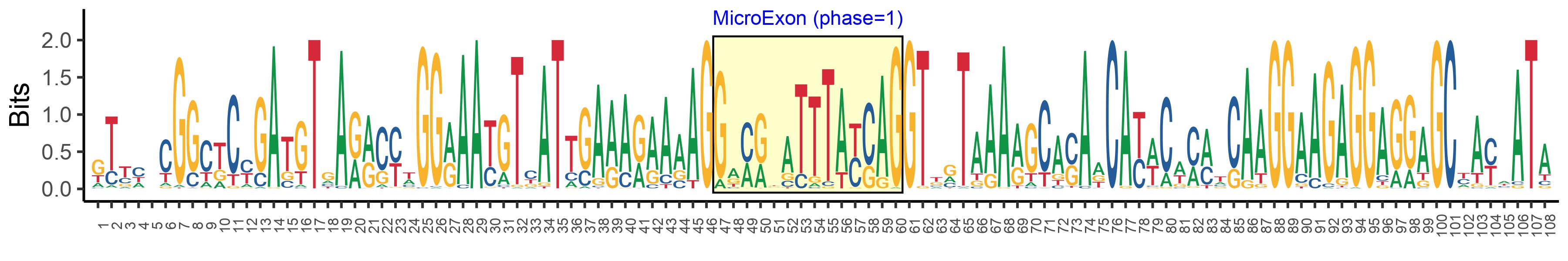 |
| Alignment of exons | 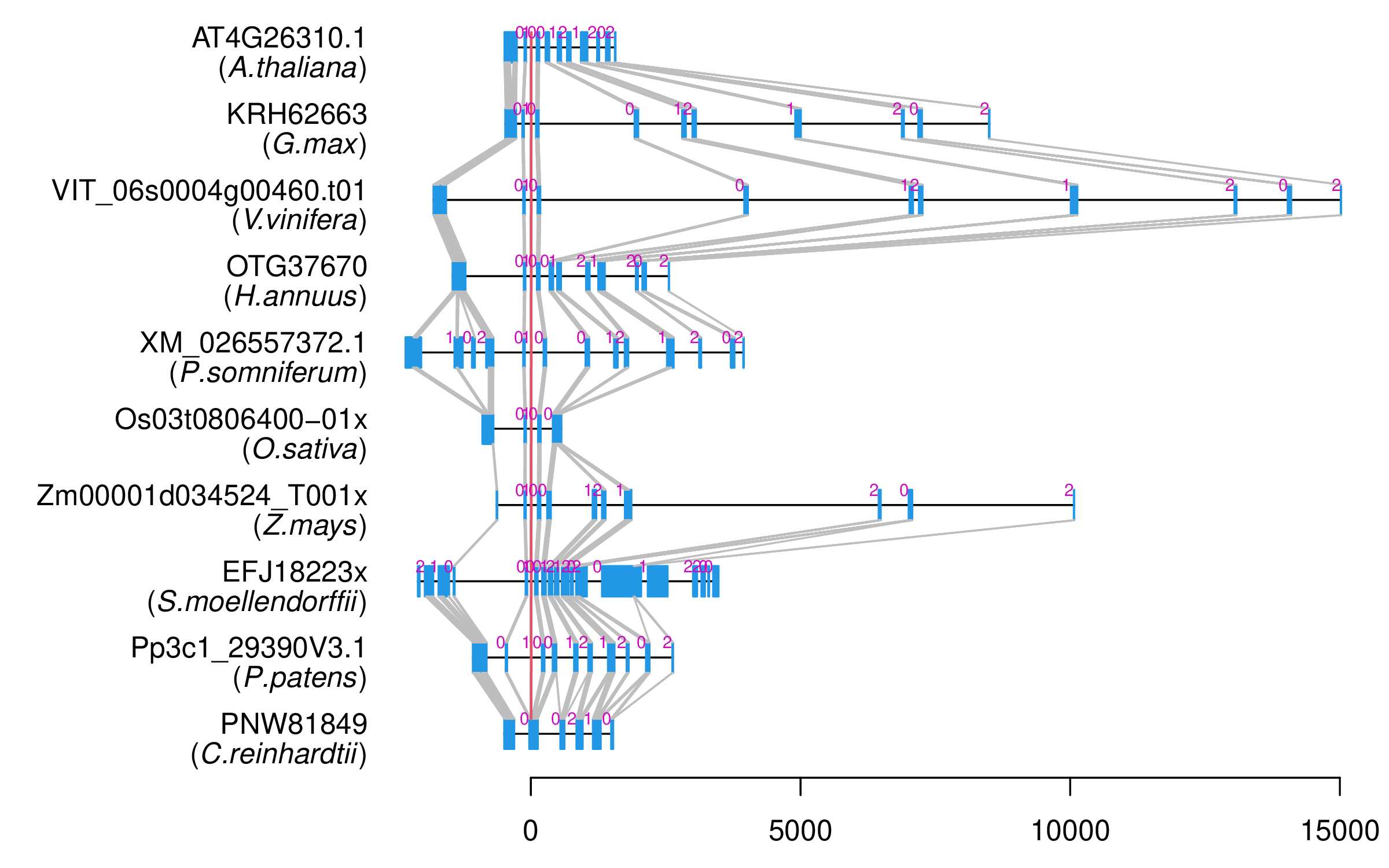 |
| Microexon DNA seq | GACGTATTTATCAG |
| Microexon Amino Acid seq | GRIYQ |
| Microexon-tag DNA Seq | GTTTTAGGCACTGATGTCAGAAGCGGGAATGTCATCCAAAGGAAAGGACGTATTTATCAGGTTCTTAAAGCACAACACACACAGCAGGGAAGAGGAGGAGCAATAATA |
| Microexon-tag Amino Acid Seq | VLGTDVRSGNVIQRKGRIYQVLKAQHTQQGRGGAII |
| Microexon-tag spanning region | 39028048-39029037 |
| Microexon-tag prediction score | 0.9472 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | XM_026569076.1x |
| Reference Transcript ID | XM_026569076.1 |
| Gene ID | NA |
| Gene Name | NA |
| Transcript ID | XM_026569076.1 |
| Protein ID | XP_026424861.1 |
| Gene ID | LOC113321197 |
| Gene Name | NA |
| Pfam domain motif | EFP_N |
| Motif E-value | 5.5e-18 |
| Motif start | 107 |
| Motif end | 161 |
| Protein seq | >XP_026424861.1 MRMKGSLTNIFRRRYRAALQDLERSTEVLKSEHKHQHTKIPHYLNIKELGLLGNSVYKGPFYRSITTLTASLQNYQTISP NNSSLSCGTFDKPWSASQHRGVKVLGTDVRSGNVIQRKGRIYQVLKAQHTQQGRGGAIIQVELRDVDSGNKVTERFRTDE AMERVFVEEKSFTYLYTIGESVTLMEPETYEQMEVTKDLFGKASAYLKEDMKVSLQLYDGKVMSATIPPRVTCTVAEAKG NMKGLTVTPQYRRVLLENGLTVQAPPFIEVGDQIIVNTVDDSYVTRAKE* |
| CDS seq | >XM_026569076.1 ATGAGGATGAAAGGGAGTTTGACTAATATCTTCAGACGAAGATATCGTGCAGCTCTTCAAGATTTAGAAAGATCAACAGA AGTTTTAAAGTCCGAACACAAACACCAGCACACTAAGATTCCTCATTATCTGAACATAAAAGAACTTGGTTTACTGGGAA ATTCAGTTTACAAAGGACCTTTTTACAGAAGCATCACAACTCTCACGGCATCACTACAAAATTACCAGACCATCTCACCT AATAATAGTTCGCTTTCGTGTGGTACCTTTGACAAACCATGGTCCGCTTCCCAACACCGTGGAGTTAAAGTTTTAGGCAC TGATGTCAGAAGCGGGAATGTCATCCAAAGGAAAGGACGTATTTATCAGGTTCTTAAAGCACAACACACACAGCAGGGAA GAGGAGGAGCAATAATACAGGTGGAATTACGTGATGTTGACAGTGGAAACAAAGTGACCGAAAGATTTCGCACCGATGAG GCTATGGAAAGAGTTTTTGTCGAAGAGAAATCTTTCACCTATTTGTACACCATAGGCGAGTCAGTAACACTAATGGAGCC TGAAACATATGAACAAATGGAGGTAACGAAGGACTTGTTTGGCAAGGCTTCTGCATACCTAAAAGAGGATATGAAAGTCA GCTTGCAATTATATGATGGTAAAGTGATGTCTGCTACTATTCCACCACGGGTGACATGTACTGTTGCTGAAGCAAAAGGC AATATGAAGGGGCTGACTGTCACACCTCAGTATAGAAGAGTTTTGCTGGAAAATGGTCTTACAGTGCAAGCGCCTCCATT CATTGAAGTTGGTGATCAGATAATTGTCAATACAGTCGATGACTCATATGTCACTAGGGCGAAGGAGTGA |
| Microexon DNA seq | GACGTATTTATCAG |
| Microexon Amino Acid seq | GRIYQ |
| Microexon-tag DNA Seq | GTTTTAGGCACTGATGTCAGAAGCGGGAATGTCATCCAAAGGAAAGGACGTATTTATCAGGTTCTTAAAGCACAACACACACAGCAGGGAAGAGGAGGAGCAATAATA |
| Microexon-tag Amino Acid seq | VLGTDVRSGNVIQRKGRIYQVLKAQHTQQGRGGAII |
| Transcript ID | XM_026569076.1 |
| Gene ID | Ps.65337 |
| Gene Name | NA |
| Pfam domain motif | EFP_N |
| Motif E-value | 5.6e-18 |
| Motif start | 107 |
| Motif end | 161 |
| Protein seq | >XM_026569076.1 MRMKGSLTNIFRRRYRAALQDLERSTEVLKSEHKHQHTKIPHYLNIKELGLLGNSVYKGPFYRSITTLTASLQNYQTISP NNSSLSCGTFDKPWSASQHRGVKVLGTDVRSGNVIQRKGRIYQVLKAQHTQQGRGGAIIQVELRDVDSGNKVTERFRTDE AMERVFVEEKSFTYLYTIGESVTLMEPETYEQMEVTKDLFGKASAYLKEDMKVSLQLYDGKVMSATIPPRVTCTVAEAKG NMKGLTVTPQYRRVLLENGLTVQAPPFIEVGDQIIVNTVDDSYVTRAKE* |
| CDS seq | >XM_026569076.1 ATGAGGATGAAAGGGAGTTTGACTAATATCTTCAGACGAAGATATCGTGCAGCTCTTCAAGATTTAGAAAGATCAACAGA AGTTTTAAAGTCCGAACACAAACACCAGCACACTAAGATTCCTCATTATCTGAACATAAAAGAACTTGGTTTACTGGGAA ATTCAGTTTACAAAGGACCTTTTTACAGAAGCATCACAACTCTCACGGCATCACTACAAAATTACCAGACCATCTCACCT AATAATAGTTCGCTTTCGTGTGGTACCTTTGACAAACCATGGTCCGCTTCCCAACACCGTGGAGTTAAAGTTTTAGGCAC TGATGTCAGAAGCGGGAATGTCATCCAAAGGAAAGGACGTATTTATCAGGTTCTTAAAGCACAACACACACAGCAGGGAA GAGGAGGAGCAATAATACAGGTGGAATTACGTGATGTTGACAGTGGAAACAAAGTGACCGAAAGATTTCGCACCGATGAG GCTATGGAAAGAGTTTTTGTCGAAGAGAAATCTTTCACCTATTTGTACACCATAGGCGAGTCAGTAACACTAATGGAGCC TGAAACATATGAACAAATGGAGGTAACGAAGGACTTGTTTGGCAAGGCTTCTGCATACCTAAAAGAGGATATGAAAGTCA GCTTGCAATTATATGATGGTAAAGTGATGTCTGCTACTATTCCACCACGGGTGACATGTACTGTTGCTGAAGCAAAAGGC AATATGAAGGGGCTGACTGTCACACCTCAGTATAGAAGAGTTTTGCTGGAAAATGGTCTTACAGTGCAAGCGCCTCCATT CATTGAAGTTGGTGATCAGATAATTGTCAATACAGTCGATGACTCATATGTCACTAGGGCGAAGGAGTGA |