
| Microexon ID | Pp_1:21038827-21038840:+ |
| Species | Physcomitrium patens | Coordinates | 1:21038827..21038840 |
| Microexon Cluster ID | MEP36 |
| Size | 14 |
| Phase | 1 |
| Pfam Domain Motif | EFP_N |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 15,31,14,48 |
| Microexon location in the Microexon-tag | 3 |
| Microexon-tag DNA Seq | KTYYGCGGCTCYGATGTGARRCYWGGRAAYGTCATTGAAARAAAAGGAMRBATTTAYSAGGTKRTAAARGCASAACAYWCWMMTCAAGGAAGAGGAGGAGCYAYWATA |
| Logo of Microexon-tag DNA Seq | 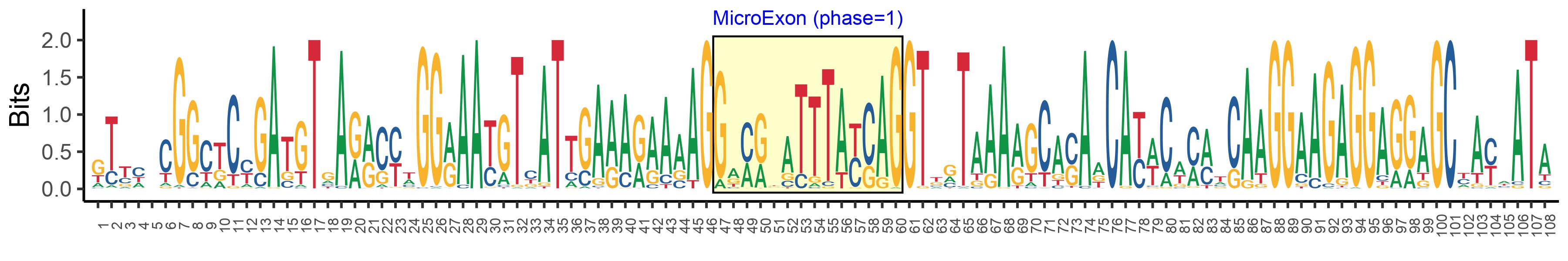 |
| Alignment of exons | 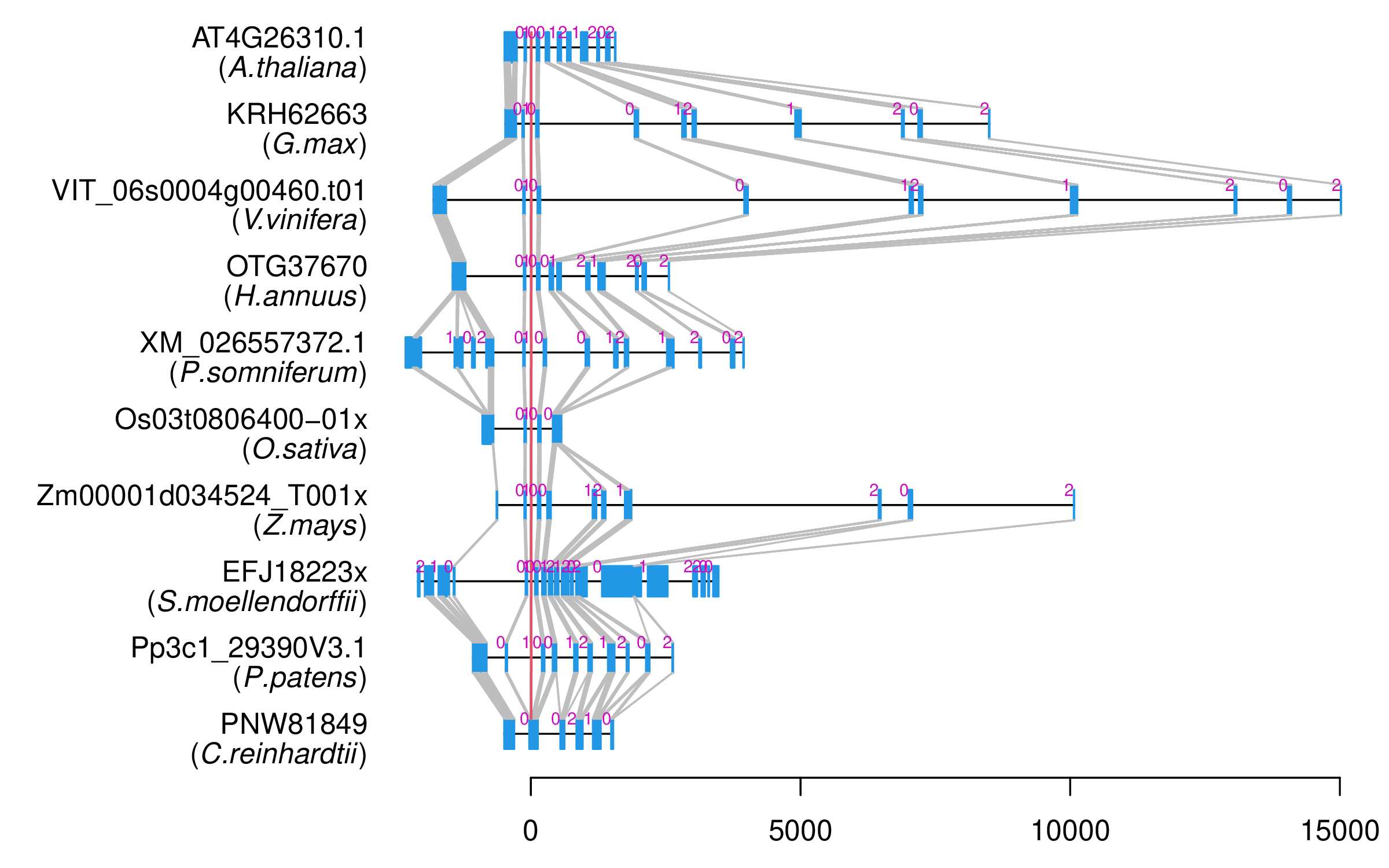 |
| Microexon DNA seq | GACGCATTTATCAG |
| Microexon Amino Acid seq | GRIYQ |
| Microexon-tag DNA Seq | GTGAACGGCTCAGAGGTCAAGCAAGGGAATGTCATTGAGCGCAAAGGACGCATTTATCAGGTACTGAAAACTCAACATACCCAACAAGGTCGAGGTGGAGCAACTATC |
| Microexon-tag Amino Acid Seq | VNGSEVKQGNVIERKGRIYQVLKTQHTQQGRGGATI |
| Microexon-tag spanning region | 21037994-21039077 |
| Microexon-tag prediction score | 0.9232 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | Pp3c1_29390V3.1x |
| Reference Transcript ID | Pp3c1_29390V3.1 |
| Gene ID | Pp3c1_29390 |
| Gene Name | NA |
| Transcript ID | Pp3c1_29390V3.1 |
| Protein ID | Pp3c1_29390V3.1 |
| Gene ID | Pp3c1_29390 |
| Gene Name | NA |
| Pfam domain motif | EFP_N |
| Motif E-value | 3.8e-17 |
| Motif start | 84 |
| Motif end | 139 |
| Protein seq | >Pp3c1_29390V3.1 MMQRYRLLARAVQRGAREYGLPPSSSPQIAPGFLRSPSNLQLWRALWSLVRDGMSESRNCFAICSDALRGGGLRQQVRHA KVNGSEVKQGNVIERKGRIYQVLKTQHTQQGRGGATIQVELRDVQSGLKLTERFRTSESIERVFVDEKAYTFLYMEGSSV VLMDPKTFDQLELSKDMLGSGAAYLSDGMEVMVQQYNGQPFSATVPPKVTCTVAEAEPYFKGQSATPTYKRIILENGQTI LAPSFITAGDQVVIDTAENTYITRSKEK* |
| CDS seq | >Pp3c1_29390V3.1 ATGATGCAGCGCTACAGATTGTTGGCGCGGGCTGTACAACGAGGTGCGCGAGAGTATGGGCTACCTCCCTCGTCGTCTCC GCAGATTGCACCAGGATTTTTGCGATCTCCATCCAATTTGCAATTATGGCGGGCGCTTTGGAGTCTTGTGAGGGACGGTA TGTCAGAATCGAGGAATTGTTTTGCTATCTGCAGCGACGCTTTGAGGGGCGGAGGCTTGAGGCAGCAAGTTCGACACGCG AAAGTGAACGGCTCAGAGGTCAAGCAAGGGAATGTCATTGAGCGCAAAGGACGCATTTATCAGGTACTGAAAACTCAACA TACCCAACAAGGTCGAGGTGGAGCAACTATCCAGGTGGAGCTTCGGGACGTGCAAAGTGGGCTGAAATTAACTGAAAGAT TCCGAACATCAGAATCTATTGAAAGAGTGTTCGTGGACGAGAAAGCCTACACTTTCTTGTACATGGAAGGTTCTAGTGTT GTTTTGATGGACCCAAAAACTTTTGATCAATTAGAACTCTCGAAGGACATGCTAGGTAGTGGAGCTGCCTACCTATCTGA TGGAATGGAGGTGATGGTTCAGCAATACAACGGCCAGCCTTTTTCCGCGACTGTTCCACCGAAGGTCACTTGCACTGTTG CAGAAGCAGAGCCATACTTCAAAGGCCAATCCGCAACACCTACGTACAAGCGCATTATATTGGAGAATGGTCAGACGATT CTGGCACCGTCATTTATTACTGCAGGAGATCAAGTAGTTATTGATACAGCGGAGAATACCTACATAACTAGAAGCAAGGA AAAATAG |
| Microexon DNA seq | GACGCATTTATCAG |
| Microexon Amino Acid seq | GRIYQ |
| Microexon-tag DNA Seq | GTGAACGGCTCAGAGGTCAAGCAAGGGAATGTCATTGAGCGCAAAGGACGCATTTATCAGGTACTGAAAACTCAACATACCCAACAAGGTCGAGGTGGAGCAACTATC |
| Microexon-tag Amino Acid seq | VNGSEVKQGNVIERKGRIYQVLKTQHTQQGRGGATI |
| Transcript ID | Pp3c1_29390V3.2 |
| Gene ID | Pp.1125 |
| Gene Name | NA |
| Pfam domain motif | EFP_N |
| Motif E-value | 3.9e-17 |
| Motif start | 84 |
| Motif end | 139 |
| Protein seq | >Pp3c1_29390V3.2 MMQRYRLLARAVQRGAREYGLPPSSSPQIAPGFLRSPSNLQLWRALWSLVRDGMSESRNCFAICSDALRGGGLRQQVRHA KVNGSEVKQGNVIERKGRIYQVLKTQHTQQGRGGATIQVELRDVQSGLKLTERFRTSESIERVFVDEKAYTFLYMEGSSV VLMDPKTFDQLELSKDMLGSGAAYLSDGMEVMVQQYNGQPFSATVPPKVTCTVAEAEPYFKGQSATPTYKRIILENGQTI LAPSFITAGDQVVIDTAENTYITRCVFRPSILR* |
| CDS seq | >Pp3c1_29390V3.2 ATGATGCAGCGCTACAGATTGTTGGCGCGGGCTGTACAACGAGGTGCGCGAGAGTATGGGCTACCTCCCTCGTCGTCTCC GCAGATTGCACCAGGATTTTTGCGATCTCCATCCAATTTGCAATTATGGCGGGCGCTTTGGAGTCTTGTGAGGGACGGTA TGTCAGAATCGAGGAATTGTTTTGCTATCTGCAGCGACGCTTTGAGGGGCGGAGGCTTGAGGCAGCAAGTTCGACACGCG AAAGTGAACGGCTCAGAGGTCAAGCAAGGGAATGTCATTGAGCGCAAAGGACGCATTTATCAGGTACTGAAAACTCAACA TACCCAACAAGGTCGAGGTGGAGCAACTATCCAGGTGGAGCTTCGGGACGTGCAAAGTGGGCTGAAATTAACTGAAAGAT TCCGAACATCAGAATCTATTGAAAGAGTGTTCGTGGACGAGAAAGCCTACACTTTCTTGTACATGGAAGGTTCTAGTGTT GTTTTGATGGACCCAAAAACTTTTGATCAATTAGAACTCTCGAAGGACATGCTAGGTAGTGGAGCTGCCTACCTATCTGA TGGAATGGAGGTGATGGTTCAGCAATACAACGGCCAGCCTTTTTCCGCGACTGTTCCACCGAAGGTCACTTGCACTGTTG CAGAAGCAGAGCCATACTTCAAAGGCCAATCCGCAACACCTACGTACAAGCGCATTATATTGGAGAATGGTCAGACGATT CTGGCACCGTCATTTATTACTGCAGGAGATCAAGTAGTTATTGATACAGCGGAGAATACCTACATAACTAGGTGCGTGTT CAGGCCATCCATTCTGCGCTAA |