
| Microexon ID | Zm_5:132599863-132599876:+ |
| Species | Zea mays | Coordinates | 5:132599863..132599876 |
| Microexon Cluster ID | MEP35 |
| Size | 14 |
| Phase | 0 |
| Pfam Domain Motif | Tudor-knot |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,14,46 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | AARGCTGARHTYCRRAAGAAKGARTGGARATAYTTYGTYCATTAYCTTGGTTGGAAYAAAAATTGGGATGAATGGGTWGGYRYKGAYCGYYTGWTGAARYWYACTGAR |
| Logo of Microexon-tag DNA Seq | 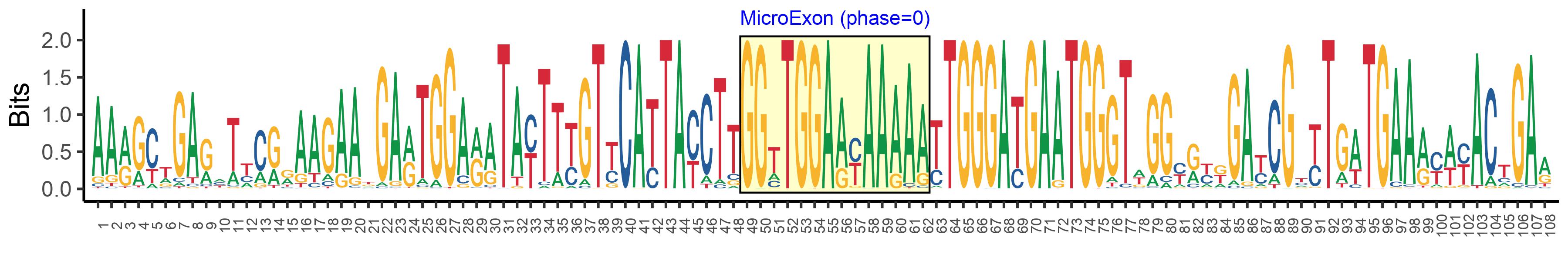 |
| Alignment of exons | 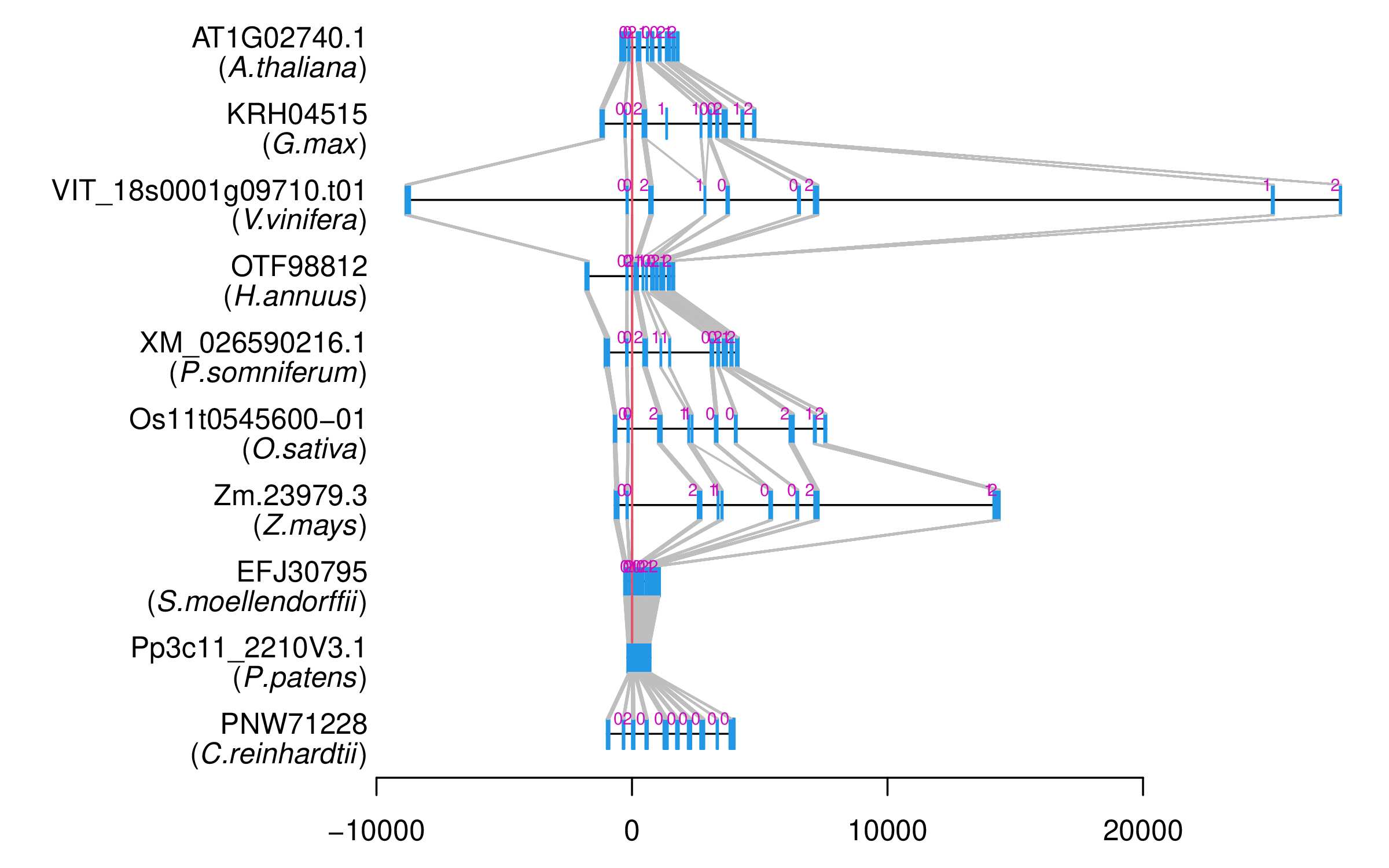 |
| Microexon DNA seq | GGTTGGAATAAGAA |
| Microexon Amino Acid seq | GWNKN |
| Microexon-tag DNA Seq | AGGATCGAAAACCATGAAGATGAATGGCGCTACTTTGTCCATTATCTCGGTTGGAATAAGAACTGGGATGAATGGGTTGCAAATGACCGCTTGTTGGAATTGACGGAG |
| Microexon-tag Amino Acid Seq | RIENHEDEWRYFVHYLGWNKNWDEWVANDRLLELTE |
| Microexon-tag spanning region | 132599655-132602484 |
| Microexon-tag prediction score | 0.9117 |
| Overlapped with the annotated transcript (%) | 57.41 |
| New Transcript ID | Zm00001d015932_T002x |
| Reference Transcript ID | Zm00001d015932_T002 |
| Gene ID | Zm00001d015932 |
| Gene Name | NA |
| Transcript ID | Zm00001d015932_T001 |
| Protein ID | Zm00001d015932_P001 |
| Gene ID | Zm00001d015932 |
| Gene Name | NA |
| Pfam domain motif | Unknown |
| Motif E-value | NA |
| Motif start | NA |
| Motif end | NA |
| Protein seq | >Zm00001d015932_P001 MGSSSNTNTSGGASDKEEKKKEDKIKGKDSSGPSFKENERVLAYHGPLLYEAKVQRIENHEDEWRYFVHYLVSLRVGIRT DAKADKDDTKSLISVKGKKRKSQLGTEIQDKEKRSSHSLLVLQFPLPLKKQLVDDWEFVTQMGKLVKLPRSPTVDDILKK YLEHRAKKDGKINDSYAEILKGLRCYFDKALPAMLLYKKERDQYAEEVKGDVSPSTVYGAEHLLRLFVKLPELLASVNME EDALNKLQLKLLDVLKFLQKNQITFFTSAYDGSCKGADEDKTK* |
| CDS seq | >Zm00001d015932_T001 ATGGGCAGCTCCTCGAATACCAACACGAGCGGCGGGGCCAGCGACAAGGAGGAGAAGAAGAAGGAGGACAAGATCAAAGG CAAGGACTCGTCGGGGCCATCCTTCAAGGAGAACGAAAGGGTCCTCGCCTACCACGGCCCCCTCCTCTACGAGGCCAAGG TTCAAAGGATCGAAAACCATGAAGATGAATGGCGCTACTTTGTCCATTATCTCGTAAGTCTTAGGGTTGGAATAAGAACG GATGCAAAAGCTGATAAAGATGACACTAAGAGTCTCATTTCAGTGAAGGGGAAGAAACGTAAAAGTCAGCTTGGGACTGA GATTCAGGACAAGGAAAAAAGATCATCACATAGTCTACTAGTGTTGCAGTTTCCTTTGCCACTGAAGAAGCAACTTGTGG ATGATTGGGAGTTTGTTACGCAGATGGGTAAGCTTGTGAAGCTACCACGATCACCTACTGTTGATGATATTCTAAAGAAG TACTTGGAACACCGGGCAAAGAAGGATGGCAAGATAAATGACTCCTATGCTGAGATTCTGAAAGGATTACGCTGCTACTT TGATAAAGCATTGCCTGCAATGCTTTTATATAAGAAAGAGCGAGATCAGTATGCTGAAGAAGTTAAAGGTGATGTCTCAC CATCAACTGTATATGGAGCTGAGCATCTATTGCGCCTTTTTGTAAAATTGCCAGAGCTACTTGCTTCTGTCAATATGGAG GAAGATGCGTTGAACAAGCTACAGCTGAAACTGCTGGACGTTCTCAAGTTTCTTCAGAAGAATCAGATCACCTTCTTTAC CTCTGCGTACGATGGTTCTTGTAAAGGTGCAGATGAGGATAAAACCAAATGA |
| Microexon DNA seq | GGTTGGAATAAGAA |
| Microexon Amino Acid seq | RVGIRT |
| Microexon-tag DNA Seq | CATGAAGATGAATGGCGCTACTTTGTCCATTATCTCGTAAGTCTTAGGGTTGGAATAAGAACGGATGCAAAAGCTGATAAAGATGACACTAAGAGTCTCATTTCAGTG |
| Microexon-tag Amino Acid seq | HEDEWRYFVHYLVSLRVGIRTDAKADKDDTKSLISV |
| Transcript ID | Zm00001d015932_T001 |
| Gene ID | Zm.23979 |
| Gene Name | NA |
| Pfam domain motif | Unknown |
| Motif E-value | NA |
| Motif start | NA |
| Motif end | NA |
| Protein seq | >Zm00001d015932_T001 MGSSSNTNTSGGASDKEEKKKEDKIKGKDSSGPSFKENERVLAYHGPLLYEAKVQRIENHEDEWRYFVHYLVSLRVGIRT DAKADKDDTKSLISVKGKKRKSQLGTEIQDKEKRSSHSLLVLQFPLPLKKQLVDDWEFVTQMGKLVKLPRSPTVDDILKK YLEHRAKKDGKINDSYAEILKGLRCYFDKALPAMLLYKKERDQYAEEVKGDVSPSTVYGAEHLLRLFVKLPELLASVNME EDALNKLQLKLLDVLKFLQKNQITFFTSAYDGSCKGADEDKTK* |
| CDS seq | >Zm00001d015932_T001 ATGGGCAGCTCCTCGAATACCAACACGAGCGGCGGGGCCAGCGACAAGGAGGAGAAGAAGAAGGAGGACAAGATCAAAGG CAAGGACTCGTCGGGGCCATCCTTCAAGGAGAACGAAAGGGTCCTCGCCTACCACGGCCCCCTCCTCTACGAGGCCAAGG TTCAAAGGATCGAAAACCATGAAGATGAATGGCGCTACTTTGTCCATTATCTCGTAAGTCTTAGGGTTGGAATAAGAACG GATGCAAAAGCTGATAAAGATGACACTAAGAGTCTCATTTCAGTGAAGGGGAAGAAACGTAAAAGTCAGCTTGGGACTGA GATTCAGGACAAGGAAAAAAGATCATCACATAGTCTACTAGTGTTGCAGTTTCCTTTGCCACTGAAGAAGCAACTTGTGG ATGATTGGGAGTTTGTTACGCAGATGGGTAAGCTTGTGAAGCTACCACGATCACCTACTGTTGATGATATTCTAAAGAAG TACTTGGAACACCGGGCAAAGAAGGATGGCAAGATAAATGACTCCTATGCTGAGATTCTGAAAGGATTACGCTGCTACTT TGATAAAGCATTGCCTGCAATGCTTTTATATAAGAAAGAGCGAGATCAGTATGCTGAAGAAGTTAAAGGTGATGTCTCAC CATCAACTGTATATGGAGCTGAGCATCTATTGCGCCTTTTTGTAAAATTGCCAGAGCTACTTGCTTCTGTCAATATGGAG GAAGATGCGTTGAACAAGCTACAGCTGAAACTGCTGGACGTTCTCAAGTTTCTTCAGAAGAATCAGATCACCTTCTTTAC CTCTGCGTACGATGGTTCTTGTAAAGGTGCAGATGAGGATAAAACCAAATGA |