
| Microexon ID | Vv_18:8101012-8101025:+ |
| Species | Vistis vinifera | Coordinates | 18:8101012..8101025 |
| Microexon Cluster ID | MEP35 |
| Size | 14 |
| Phase | 0 |
| Pfam Domain Motif | Tudor-knot |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,14,46 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | AARGCTGARHTYCRRAAGAAKGARTGGARATAYTTYGTYCATTAYCTTGGTTGGAAYAAAAATTGGGATGAATGGGTWGGYRYKGAYCGYYTGWTGAARYWYACTGAR |
| Logo of Microexon-tag DNA Seq | 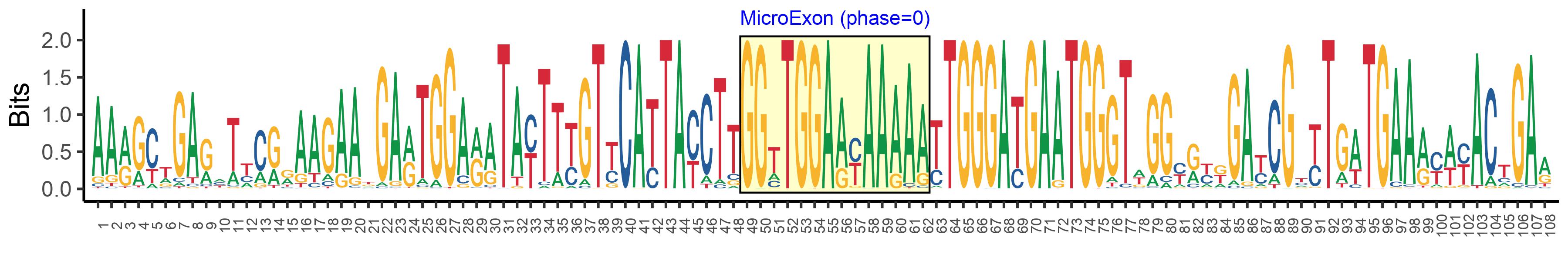 |
| Alignment of exons | 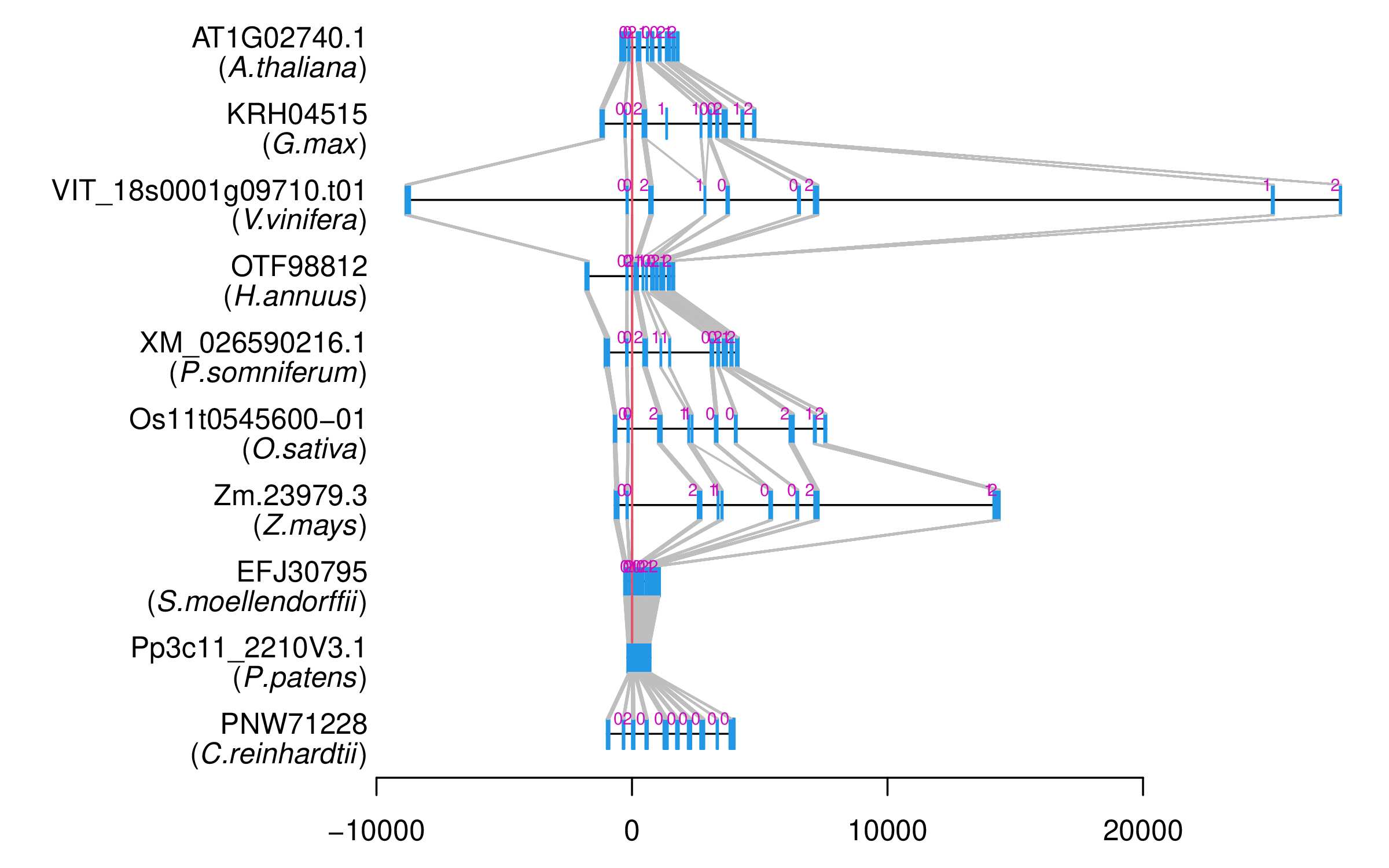 |
| Microexon DNA seq | GGTTGGAGCAAAAA |
| Microexon Amino Acid seq | GWSKN |
| Microexon-tag DNA Seq | AGAATTGAATTTCAGATGAATGAATGGAAGTATTTTGTTCATTACCTTGGTTGGAGCAAAAACTGGGATGAATGGGTGGGCATGGATCGGCTGCTGAAATTTAGTGAA |
| Microexon-tag Amino Acid Seq | RIEFQMNEWKYFVHYLGWSKNWDEWVGMDRLLKFSE |
| Microexon-tag spanning region | 8100803-8101730 |
| Microexon-tag prediction score | 0.9527 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | VIT_18s0001g09710.t01x |
| Reference Transcript ID | VIT_18s0001g09710.t01 |
| Gene ID | VIT_18s0001g09710 |
| Gene Name | NA |
| Transcript ID | VIT_18s0001g09710.t01 |
| Protein ID | VIT_18s0001g09710.t01 |
| Gene ID | VIT_18s0001g09710 |
| Gene Name | NA |
| Pfam domain motif | Tudor-knot |
| Motif E-value | 3e-09 |
| Motif start | 42 |
| Motif end | 92 |
| Protein seq | >VIT_18s0001g09710.t01 MGSSVDASATETDGDTTETDIDVDADSDTDDYPPLSDSCPFSEGEKVLAYHNLRIYPAKVRRIEFQMNEWKYFVHYLGWS KNWDEWVGMDRLLKFSEENVQKQKALGKKQGIDKNTKPVRASQIKPKNFARGKKWKNDCVTKEAIPVEKLVNIQIPPTLK KQLVDDCEFITHLGQLIRLPRAPTVDKILKKYLDYRIKRDGMISDSAGEILKGLRCYFDKALPVMLLYERERQQYQEAIA NNVSPSTIYGAEHLLRLFVKLPELLFHANIEKETSKELQMELLDFLKFLQKNQSAFFLTSYISSA* |
| CDS seq | >VIT_18s0001g09710.t01 ATGGGAAGCTCTGTCGATGCCTCCGCAACCGAAACCGACGGCGACACCACCGAAACCGACATCGACGTCGACGCCGACAG CGACACCGATGACTACCCTCCCCTTTCCGATTCGTGTCCCTTTTCTGAAGGAGAGAAGGTCCTCGCTTATCATAACCTTC GTATTTATCCAGCCAAGGTGCGAAGAATTGAATTTCAGATGAATGAATGGAAGTATTTTGTTCATTACCTTGGTTGGAGC AAAAACTGGGATGAATGGGTGGGCATGGATCGGCTGCTGAAATTTAGTGAAGAGAATGTCCAAAAACAGAAGGCCCTGGG AAAAAAACAGGGCATAGACAAAAACACAAAGCCAGTGCGTGCCTCACAGATTAAGCCAAAAAATTTTGCAAGAGGCAAGA AGTGGAAGAATGACTGTGTCACTAAGGAAGCTATACCAGTGGAGAAGCTTGTGAACATCCAAATTCCTCCAACACTGAAG AAGCAACTAGTTGATGATTGTGAATTCATTACTCATCTGGGCCAGCTTATTAGACTGCCTCGAGCTCCAACTGTTGACAA AATATTGAAGAAGTATCTTGATTACAGGATAAAGAGGGATGGCATGATATCTGATTCGGCTGGAGAAATTCTGAAAGGGT TACGTTGTTACTTTGACAAAGCATTGCCAGTGATGCTTCTGTATGAAAGGGAGCGTCAACAGTATCAAGAAGCAATTGCA AATAATGTCTCTCCTTCAACTATATATGGTGCTGAACATCTATTACGCCTCTTTGTTAAATTGCCCGAGTTATTATTCCA TGCCAACATTGAAAAGGAGACATCAAAAGAGTTACAAATGGAATTACTCGACTTTCTCAAGTTCCTGCAGAAGAATCAGA GTGCCTTCTTCCTCACCTCATATATTTCTTCCGCATGA |
| Microexon DNA seq | GGTTGGAGCAAAAA |
| Microexon Amino Acid seq | GWSKN |
| Microexon-tag DNA Seq | AGAATTGAATTTCAGATGAATGAATGGAAGTATTTTGTTCATTACCTTGGTTGGAGCAAAAACTGGGATGAATGGGTGGGCATGGATCGGCTGCTGAAATTTAGTGAA |
| Microexon-tag Amino Acid seq | RIEFQMNEWKYFVHYLGWSKNWDEWVGMDRLLKFSE |
| Transcript ID | Vv.15856.1 |
| Gene ID | Vv.15856 |
| Gene Name | NA |
| Pfam domain motif | Tudor-knot |
| Motif E-value | 3e-09 |
| Motif start | 42 |
| Motif end | 92 |
| Protein seq | >Vv.15856.1 MGSSVDASATETDGDTTETDIDVDADSDTDDYPPLSDSCPFSEGEKVLAYHNLRIYPAKVRRIEFQMNEWKYFVHYLGWS KNWDEWVGMDRLLKFSEENVQKQKALGKKQGIDKNTKPVRASQIKPKNFARGKKWKNDCVTKKEAIPVEKLVNIQIPPTL KKQLVDDCEFITHLGQLIRLPRAPTVDKILKKYLDYRIKRDGMISDSAGEILKGLRCYFDKALPVMLLYERERQQYQEAI ANNVSPSTIYGAEHLLRLFVKLPELLFHANIEKETSKELQMELLDFLKFLQKNQSAFFLTSYISSA* |
| CDS seq | >Vv.15856.1 ATGGGAAGCTCTGTCGATGCCTCCGCAACCGAAACCGACGGCGACACCACCGAAACCGACATCGACGTCGACGCCGACAG CGACACCGATGACTACCCTCCCCTTTCCGATTCGTGTCCCTTTTCTGAAGGAGAGAAGGTCCTCGCTTATCATAACCTTC GTATTTATCCAGCCAAGGTGCGAAGAATTGAATTTCAGATGAATGAATGGAAGTATTTTGTTCATTACCTTGGTTGGAGC AAAAACTGGGATGAATGGGTGGGCATGGATCGGCTGCTGAAATTTAGTGAAGAGAATGTCCAAAAACAGAAGGCCCTGGG AAAAAAACAGGGCATAGACAAAAACACAAAGCCAGTGCGTGCCTCACAGATTAAGCCAAAAAATTTTGCAAGAGGCAAGA AGTGGAAGAATGACTGTGTCACTAAGAAGGAAGCTATACCAGTGGAGAAGCTTGTGAACATCCAAATTCCTCCAACACTG AAGAAGCAACTAGTTGATGATTGTGAATTCATTACTCATCTGGGCCAGCTTATTAGACTGCCTCGAGCTCCAACTGTTGA CAAAATATTGAAGAAGTATCTTGATTACAGGATAAAGAGGGATGGCATGATATCTGATTCGGCTGGAGAAATTCTGAAAG GGTTACGTTGTTACTTTGACAAAGCATTGCCAGTGATGCTTCTGTATGAAAGGGAGCGTCAACAGTATCAAGAAGCAATT GCAAATAATGTCTCTCCTTCAACTATATATGGTGCTGAACATCTATTACGCCTCTTTGTTAAATTGCCCGAGTTATTATT CCATGCCAACATTGAAAAGGAGACATCAAAAGAGTTACAAATGGAATTACTCGACTTTCTCAAGTTCCTGCAGAAGAATC AGAGTGCCTTCTTCCTCACCTCATATATTTCTTCCGCATGA |