
| Microexon ID | Ps_NC_039360.1:97678854-97678867:- |
| Species | Papaver somniferum | Coordinates | NC_039360.1:97678854..97678867 |
| Microexon Cluster ID | MEP35 |
| Size | 14 |
| Phase | 0 |
| Pfam Domain Motif | Tudor-knot |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,14,46 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | AARGCTGARHTYCRRAAGAAKGARTGGARATAYTTYGTYCATTAYCTTGGTTGGAAYAAAAATTGGGATGAATGGGTWGGYRYKGAYCGYYTGWTGAARYWYACTGAR |
| Logo of Microexon-tag DNA Seq | 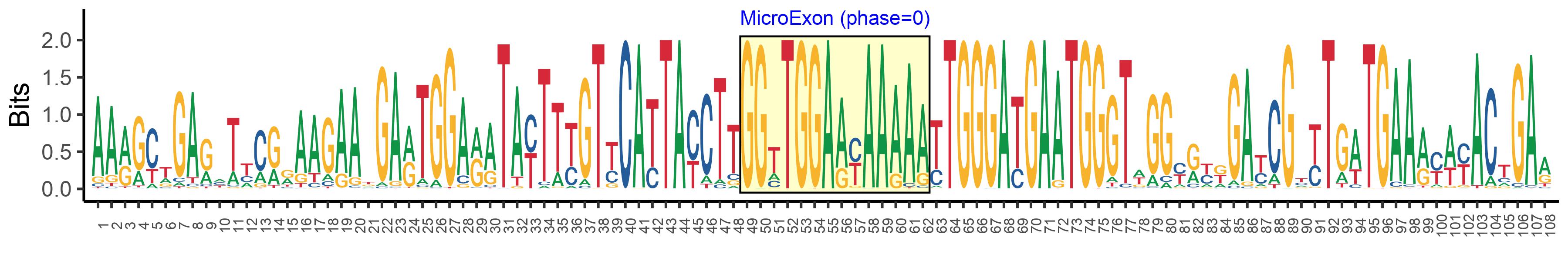 |
| Alignment of exons | 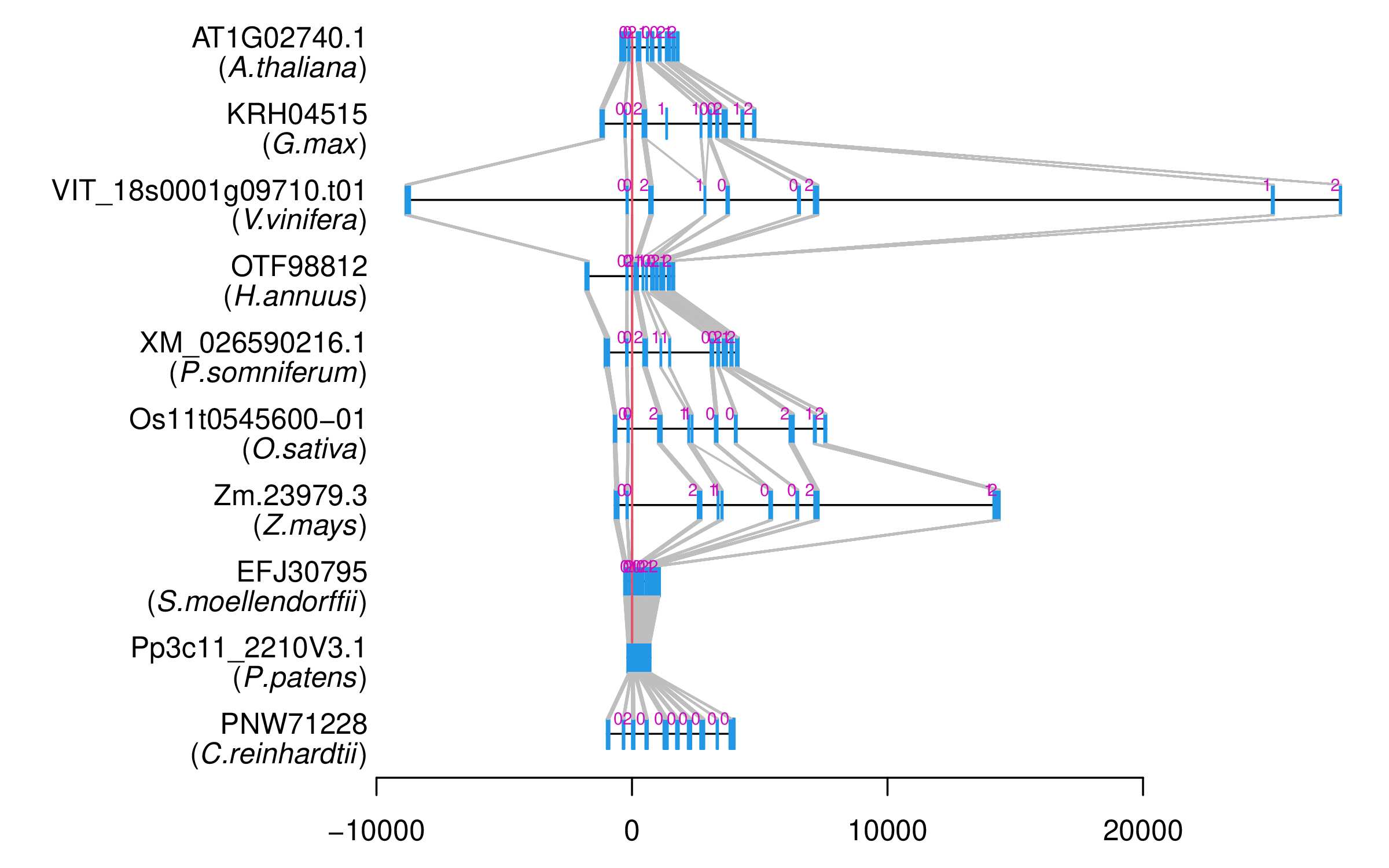 |
| Microexon DNA seq | GGCTGGAACAAAAA |
| Microexon Amino Acid seq | GWNKN |
| Microexon-tag DNA Seq | CGGAAAGTTGATACTACGGAGGAAAAGAGATACTTTGTTCATTACCTTGGCTGGAACAAAAACTGGGATGAATGGGTTGGCCCGGAACGTTTGTTGAAATTCACTGAG |
| Microexon-tag Amino Acid Seq | RKVDTTEEKRYFVHYLGWNKNWDEWVGPERLLKFTE |
| Microexon-tag spanning region | 97678378-97679063 |
| Microexon-tag prediction score | 0.918 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | XM_026600157.1x |
| Reference Transcript ID | XM_026600157.1 |
| Gene ID | NA |
| Gene Name | NA |
| Transcript ID | XM_026600157.1 |
| Protein ID | XP_026455942.1 |
| Gene ID | LOC113356920 |
| Gene Name | NA |
| Pfam domain motif | Tudor-knot |
| Motif E-value | 1.3e-09 |
| Motif start | 43 |
| Motif end | 96 |
| Protein seq | >XP_026455942.1 MGSPKSGGAKEDTDVDIVGDSPSNSSGEADPSSSSLGISAYTEGERVFAYHGPRIYESKVQKVEMRKVDTTEEKRYFVHY LGWNKNWDEWVGPERLLKFTEENKKLQEDLKLKQQGTDKNPKSGRSSHMKPKSSADAKMDKEDTRNYVGKGKKRKSDFSI EEKDTISNEKLVKIQIPSTLKKQLVDDWEFVVQLGKLVKLPRSPCVDDIMKRYLDYKLKKEGMIGESVGEILKGLRCYFD KALPVMLLYKMERQQYQEAIKDDVSPSSVYGAEHLLRLFVKLPELLSYANIEEEASTRLHTKLIDLLKFLQKNQSTFFLS AYDNPKGGVAEGSGEDSLEGDDN* |
| CDS seq | >XM_026600157.1 ATGGGTAGCCCTAAATCAGGAGGTGCTAAAGAAGACACAGACGTCGATATTGTTGGTGATTCTCCTTCAAATTCTAGTGG TGAAGCAGATCCTTCTTCTTCTTCTTTAGGTATTTCTGCATATACTGAAGGTGAAAGAGTTTTTGCTTATCACGGTCCCC GTATTTACGAGTCTAAGGTTCAAAAAGTTGAGATGCGGAAAGTTGATACTACGGAGGAAAAGAGATACTTTGTTCATTAC CTTGGCTGGAACAAAAACTGGGATGAATGGGTTGGCCCGGAACGTTTGTTGAAATTCACTGAGGAGAACAAGAAACTGCA GGAGGACCTTAAACTAAAACAGCAGGGTACAGACAAGAATCCCAAATCAGGGCGTTCATCACACATGAAGCCAAAAAGCT CTGCTGATGCAAAAATGGATAAAGAGGATACTAGAAACTATGTTGGGAAGGGAAAGAAGCGGAAAAGTGACTTCAGTATT GAGGAGAAGGACACCATTTCCAATGAGAAGCTTGTGAAGATCCAAATTCCATCAACACTTAAAAAACAACTGGTTGATGA TTGGGAGTTTGTAGTTCAGCTCGGCAAGCTTGTTAAGCTTCCGCGCTCTCCTTGTGTGGATGATATAATGAAGAGGTACC TTGATTACAAATTAAAGAAGGAAGGCATGATAGGCGAATCGGTTGGGGAAATCTTGAAGGGTTTGCGCTGTTACTTTGAC AAAGCATTGCCAGTCATGCTCTTGTACAAAATGGAACGCCAACAGTATCAAGAAGCAATTAAAGACGATGTTTCTCCATC TAGTGTATATGGCGCTGAGCATTTACTACGCCTATTTGTGAAGTTGCCTGAGCTACTGTCTTATGCAAACATTGAAGAGG AAGCGTCAACGCGGTTACATACTAAACTAATCGACTTACTCAAATTCTTGCAGAAAAATCAGAGCACCTTCTTTCTTTCT GCATATGATAATCCTAAAGGTGGAGTAGCAGAAGGAAGCGGTGAGGACAGTTTAGAAGGTGACGACAACTGA |
| Microexon DNA seq | GGCTGGAACAAAAA |
| Microexon Amino Acid seq | GWNKN |
| Microexon-tag DNA Seq | CGGAAAGTTGATACTACGGAGGAAAAGAGATACTTTGTTCATTACCTTGGCTGGAACAAAAACTGGGATGAATGGGTTGGCCCGGAACGTTTGTTGAAATTCACTGAG |
| Microexon-tag Amino Acid seq | RKVDTTEEKRYFVHYLGWNKNWDEWVGPERLLKFTE |
| Transcript ID | Ps.18500.1 |
| Gene ID | Ps.18500 |
| Gene Name | NA |
| Pfam domain motif | Tudor-knot |
| Motif E-value | 1.2e-09 |
| Motif start | 43 |
| Motif end | 96 |
| Protein seq | >Ps.18500.1 MGSPKSGGAKEDTDVDIVGDSPSNSSGEADPSSSSLGISAYTEGERVFAYHGPRIYESKVQKVEMRKVDTTEEKRYFVHY LGWNKNWDEWVGPERLLKFTEENKKLQEDLKLKQQGTDKNPKSGRSSHMKPKSSAVGKGKKRKSDFSIEEKDTISNEKLV KIQIPSTLKKQLVDDWEFVVQLGKLVKLPRSPCVDDIMKRIGESVGEILKGLRCYFDKALPVMLLYKMERQQYQEAIKDD VSPSSVYGAEHLLRLFVKLPELLSYANIEEEASTRLHTKLIDLLKFLQKNQSTFFLSAYDNPKGGVAEGSGEDSLEGDDN * |
| CDS seq | >Ps.18500.1 ATGGGTAGCCCTAAATCAGGAGGTGCTAAAGAAGACACAGACGTCGATATTGTTGGTGATTCTCCTTCAAATTCTAGTGG TGAAGCAGATCCTTCTTCTTCTTCTTTAGGTATTTCTGCATATACTGAAGGTGAAAGAGTTTTTGCTTATCACGGTCCCC GTATTTACGAGTCTAAGGTTCAAAAAGTTGAGATGCGGAAAGTTGATACTACGGAGGAAAAGAGATACTTTGTTCATTAC CTTGGCTGGAACAAAAACTGGGATGAATGGGTTGGCCCGGAACGTTTGTTGAAATTCACTGAGGAGAACAAGAAACTGCA GGAGGACCTTAAACTAAAACAGCAGGGTACAGACAAGAATCCCAAATCAGGGCGTTCATCACACATGAAGCCAAAAAGCT CTGCTGTTGGGAAGGGAAAGAAGCGGAAAAGTGACTTCAGTATTGAGGAGAAGGACACCATTTCCAATGAGAAGCTTGTG AAGATCCAAATTCCATCAACACTTAAAAAACAACTGGTTGATGATTGGGAGTTTGTAGTTCAGCTCGGCAAGCTTGTTAA GCTTCCGCGCTCTCCTTGTGTGGATGATATAATGAAGAGGATAGGCGAATCGGTTGGGGAAATCTTGAAGGGTTTGCGCT GTTACTTTGACAAAGCATTGCCAGTCATGCTCTTGTACAAAATGGAACGCCAACAGTATCAAGAAGCAATTAAAGACGAT GTTTCTCCATCTAGTGTATATGGCGCTGAGCATTTACTACGCCTATTTGTGAAGTTGCCTGAGCTACTGTCTTATGCAAA CATTGAAGAGGAAGCGTCAACGCGGTTACATACTAAACTAATCGACTTACTCAAATTCTTGCAGAAAAATCAGAGCACCT TCTTTCTTTCTGCATATGATAATCCTAAAGGTGGAGTAGCAGAAGGAAGCGGTGAGGACAGTTTAGAAGGTGACGACAAC TGA |