
| Microexon ID | Gm_4:3448978-3448991:- |
| Species | Glycine max | Coordinates | 4:3448978..3448991 |
| Microexon Cluster ID | MEP35 |
| Size | 14 |
| Phase | 0 |
| Pfam Domain Motif | Tudor-knot |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,14,46 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | AARGCTGARHTYCRRAAGAAKGARTGGARATAYTTYGTYCATTAYCTTGGTTGGAAYAAAAATTGGGATGAATGGGTWGGYRYKGAYCGYYTGWTGAARYWYACTGAR |
| Logo of Microexon-tag DNA Seq | 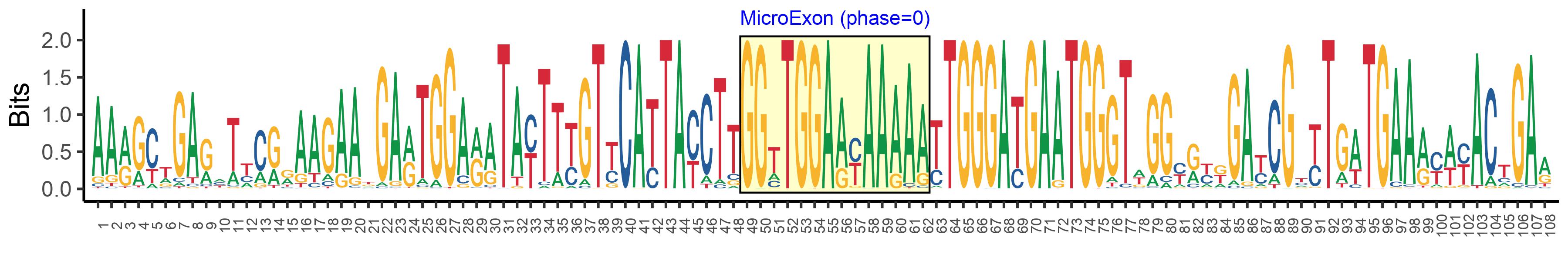 |
| Alignment of exons | 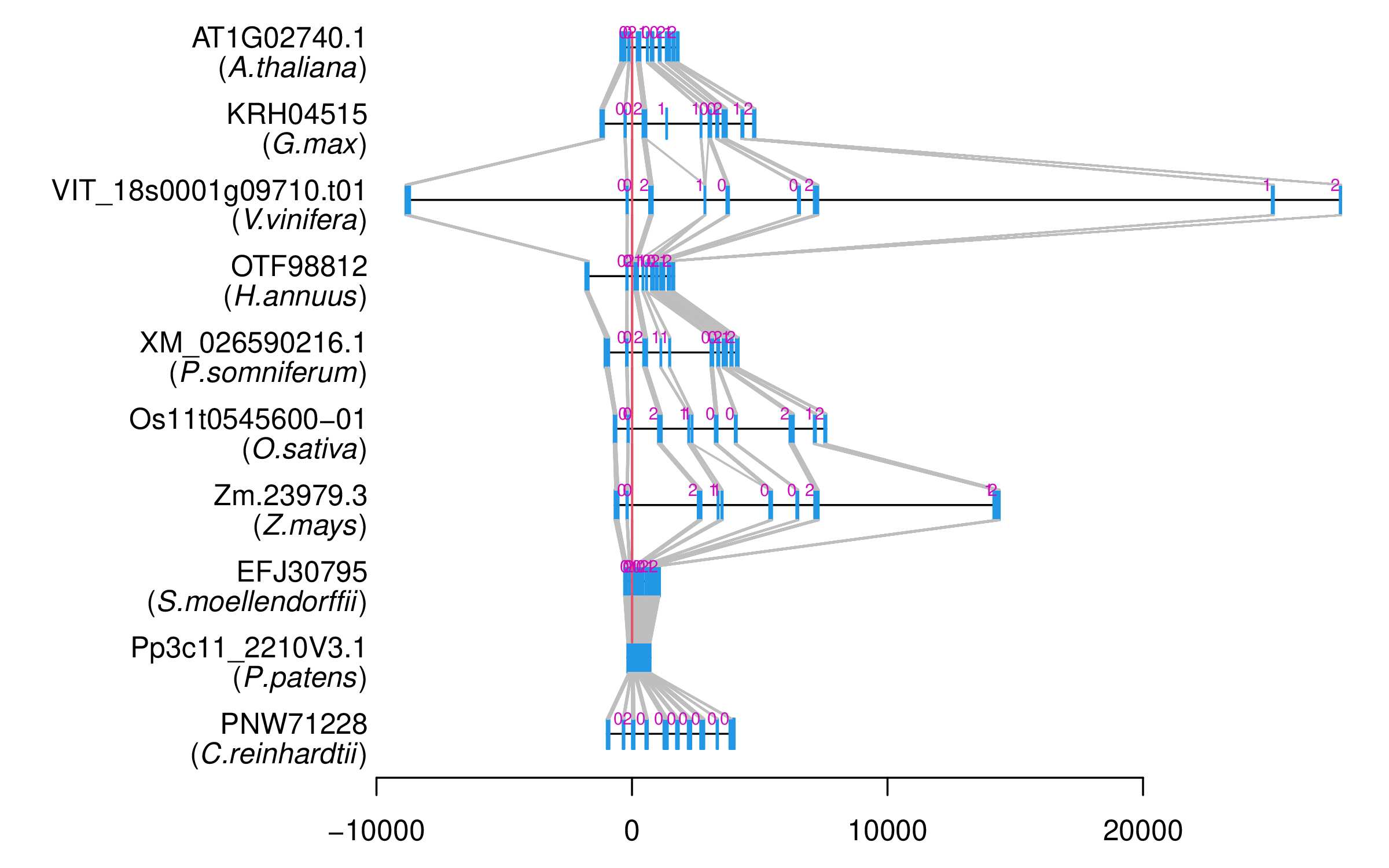 |
| Microexon DNA seq | GGTTGGAAGAAGAG |
| Microexon Amino Acid seq | GWKKR |
| Microexon-tag DNA Seq | GTTGAATACCGCAACACTAAAGGATGGCAATTCCTCGTTCACTATCTTGGTTGGAAGAAGAGATGGGATGAATGGCTTGACCTGGATCGTTTGATGAAACACACCGAG |
| Microexon-tag Amino Acid Seq | VEYRNTKGWQFLVHYLGWKKRWDEWLDLDRLMKHTE |
| Microexon-tag spanning region | 3448848-3449137 |
| Microexon-tag prediction score | 0.8839 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRH61355x |
| Reference Transcript ID | KRH61355 |
| Gene ID | GLYMA_04G042700 |
| Gene Name | NA |
| Transcript ID | KRH61355 |
| Protein ID | KRH61355 |
| Gene ID | GLYMA_04G042700 |
| Gene Name | NA |
| Pfam domain motif | Tudor-knot |
| Motif E-value | 6e-08 |
| Motif start | 41 |
| Motif end | 91 |
| Protein seq | >KRH61355 MEMGISKTTTSTEVTDDNSNSSSTTHTQTRNHISFAAPYAVGEKVLTNHSDCLYEAKVRQVEYRNTKGWQFLVHYLGWKK RWDEWLDLDRLMKHTEENMRKKHDLDEKLGNDKNAKVPRGSLAKSKPTNVSRGRKRRNESVIKGKPAVDPDKLVNIQIPP TLKKQLVDDCEFITHLGKLVKLPRTPNVKGILKNYFDYRLKKCGSVGDSVEEIMKGLSCYFDKALPVMLLYKNERQQYQE ACPANVFPSAIYGAEHLLRLFVKLPELLFHASVEEETLMELQAHLIDFLRFQCFPPGSCKKTRAHSSFLLIMLQKVLRTA PTNKCVCLVKPAYVYVWTQPVHVEELKNQLHMLLRAANFFPSQIDVIPGVLLLVSLVTITFPGYL* |
| CDS seq | >KRH61355 ATGGAAATGGGGATTTCCAAGACAACAACAAGCACTGAGGTTACCGACGACAATTCCAATAGCAGCAGCACCACTCATAC TCAAACTCGCAACCATATCTCCTTTGCTGCTCCCTACGCCGTAGGAGAAAAGGTCTTAACCAATCATTCTGATTGTCTTT ACGAAGCCAAGGTGAGGCAAGTTGAATACCGCAACACTAAAGGATGGCAATTCCTCGTTCACTATCTTGGTTGGAAGAAG AGATGGGATGAATGGCTTGACCTGGATCGTTTGATGAAACACACCGAGGAAAATATGCGCAAAAAACATGACCTTGACGA GAAGCTTGGCAATGATAAGAATGCAAAAGTCCCGCGTGGTTCTCTGGCGAAGTCCAAGCCTACTAACGTTTCAAGAGGCA GGAAACGAAGGAATGAATCTGTCATCAAGGGAAAACCTGCAGTTGATCCTGACAAGCTTGTTAACATTCAAATTCCTCCA ACACTAAAGAAACAATTAGTTGATGATTGTGAGTTTATTACCCATCTCGGCAAGCTTGTTAAACTCCCTCGTACTCCTAA TGTGAAAGGCATATTGAAAAATTATTTTGATTACAGATTGAAGAAATGTGGCTCGGTGGGTGATTCAGTTGAAGAAATTA TGAAAGGATTGTCTTGTTACTTCGACAAAGCACTGCCAGTGATGCTTTTGTACAAGAACGAGCGCCAACAGTACCAAGAG GCTTGTCCAGCCAATGTCTTTCCTTCTGCTATATATGGTGCTGAGCATTTGTTACGTCTCTTTGTTAAGCTGCCCGAGTT ACTGTTTCATGCTAGTGTTGAAGAGGAGACATTGATGGAACTTCAAGCACACTTAATTGACTTTCTCAGGTTTCAATGTT TTCCTCCAGGTTCTTGCAAAAAAACCAGAGCACATTCTTCCTTTCTACTTATCATGTTGCAGAAGGTATTGAGAACAGCA CCAACAAACAAGTGTGTTTGCCTTGTTAAGCCTGCATATGTTTATGTATGGACGCAACCTGTTCATGTGGAAGAACTCAA GAATCAATTACATATGCTTCTAAGGGCTGCAAACTTTTTTCCTAGCCAAATTGATGTCATTCCTGGAGTTCTGCTTCTGG TTAGTCTAGTGACTATCACTTTTCCTGGTTATTTATAA |
| Microexon DNA seq | GGTTGGAAGAAGAG |
| Microexon Amino Acid seq | GWKKR |
| Microexon-tag DNA Seq | GTTGAATACCGCAACACTAAAGGATGGCAATTCCTCGTTCACTATCTTGGTTGGAAGAAGAGATGGGATGAATGGCTTGACCTGGATCGTTTGATGAAACACACCGAG |
| Microexon-tag Amino Acid seq | VEYRNTKGWQFLVHYLGWKKRWDEWLDLDRLMKHTE |
| Transcript ID | Gm.37820.1 |
| Gene ID | Gm.37820 |
| Gene Name | NA |
| Pfam domain motif | Tudor-knot |
| Motif E-value | 3.6e-08 |
| Motif start | 41 |
| Motif end | 91 |
| Protein seq | >Gm.37820.1 MEMGISKTTTSTEVTDDNSNSSSTTHTQTRNHISFAAPYAVGEKVLTNHSDCLYEAKVRQVEYRNTKGWQFLVHYLGWKK RWDEWLDLDRLMKHTEENMRKKHDLDEKLGNDKNAKVPRGSLAKSKPTNVSRGRKRRNESVIKGKPAVDPDKLVNIQIPP TLKKQLVDDCEFITHLGKLVKLPRTPNVKGILKNYFDYRLKKCGSVGDSVEEIMKGLSCYFDKALPVMLLYKNERQQYQE ACPANVFPSAIYGAEHLLRLFGMLVCLPQDELII* |
| CDS seq | >Gm.37820.1 ATGGAAATGGGGATTTCCAAGACAACAACAAGCACTGAGGTTACCGACGACAATTCCAATAGCAGCAGCACCACTCATAC TCAAACTCGCAACCATATCTCCTTTGCTGCTCCCTACGCCGTAGGAGAAAAGGTCTTAACCAATCATTCTGATTGTCTTT ACGAAGCCAAGGTGAGGCAAGTTGAATACCGCAACACTAAAGGATGGCAATTCCTCGTTCACTATCTTGGTTGGAAGAAG AGATGGGATGAATGGCTTGACCTGGATCGTTTGATGAAACACACCGAGGAAAATATGCGCAAAAAACATGACCTTGACGA GAAGCTTGGCAATGATAAGAATGCAAAAGTCCCGCGTGGTTCTCTGGCGAAGTCCAAGCCTACTAACGTTTCAAGAGGCA GGAAACGAAGGAATGAATCTGTCATCAAGGGAAAACCTGCAGTTGATCCTGACAAGCTTGTTAACATTCAAATTCCTCCA ACACTAAAGAAACAATTAGTTGATGATTGTGAGTTTATTACCCATCTCGGCAAGCTTGTTAAACTCCCTCGTACTCCTAA TGTGAAAGGCATATTGAAAAATTATTTTGATTACAGATTGAAGAAATGTGGCTCGGTGGGTGATTCAGTTGAAGAAATTA TGAAAGGATTGTCTTGTTACTTCGACAAAGCACTGCCAGTGATGCTTTTGTACAAGAACGAGCGCCAACAGTACCAAGAG GCTTGTCCAGCCAATGTCTTTCCTTCTGCTATATATGGTGCTGAGCATTTGTTACGTCTCTTTGGTATGCTCGTTTGTCT GCCACAAGACGAGCTTATTATATGA |