
| Microexon ID | At_4:17548453-17548466:- |
| Species | Arabidopsis thaliana | Coordinates | 4:17548453..17548466 |
| Microexon Cluster ID | MEP35 |
| Size | 14 |
| Phase | 0 |
| Pfam Domain Motif | Tudor-knot |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,14,46 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | AARGCTGARHTYCRRAAGAAKGARTGGARATAYTTYGTYCATTAYCTTGGTTGGAAYAAAAATTGGGATGAATGGGTWGGYRYKGAYCGYYTGWTGAARYWYACTGAR |
| Logo of Microexon-tag DNA Seq | 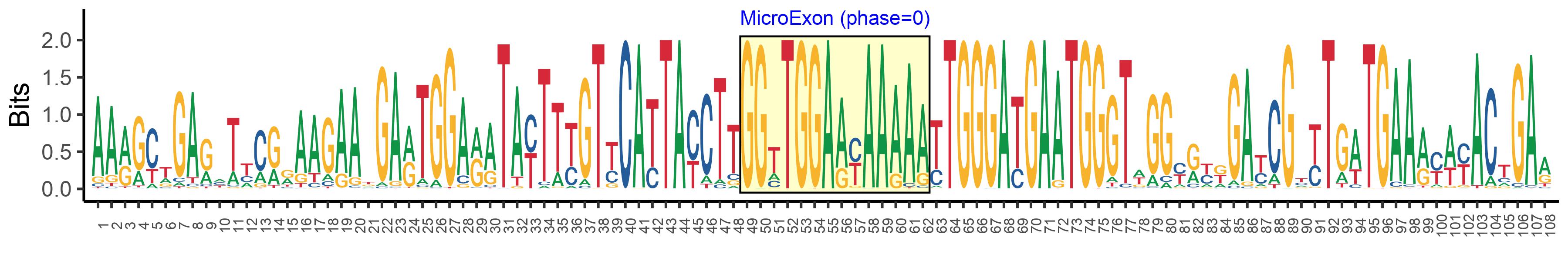 |
| Alignment of exons | 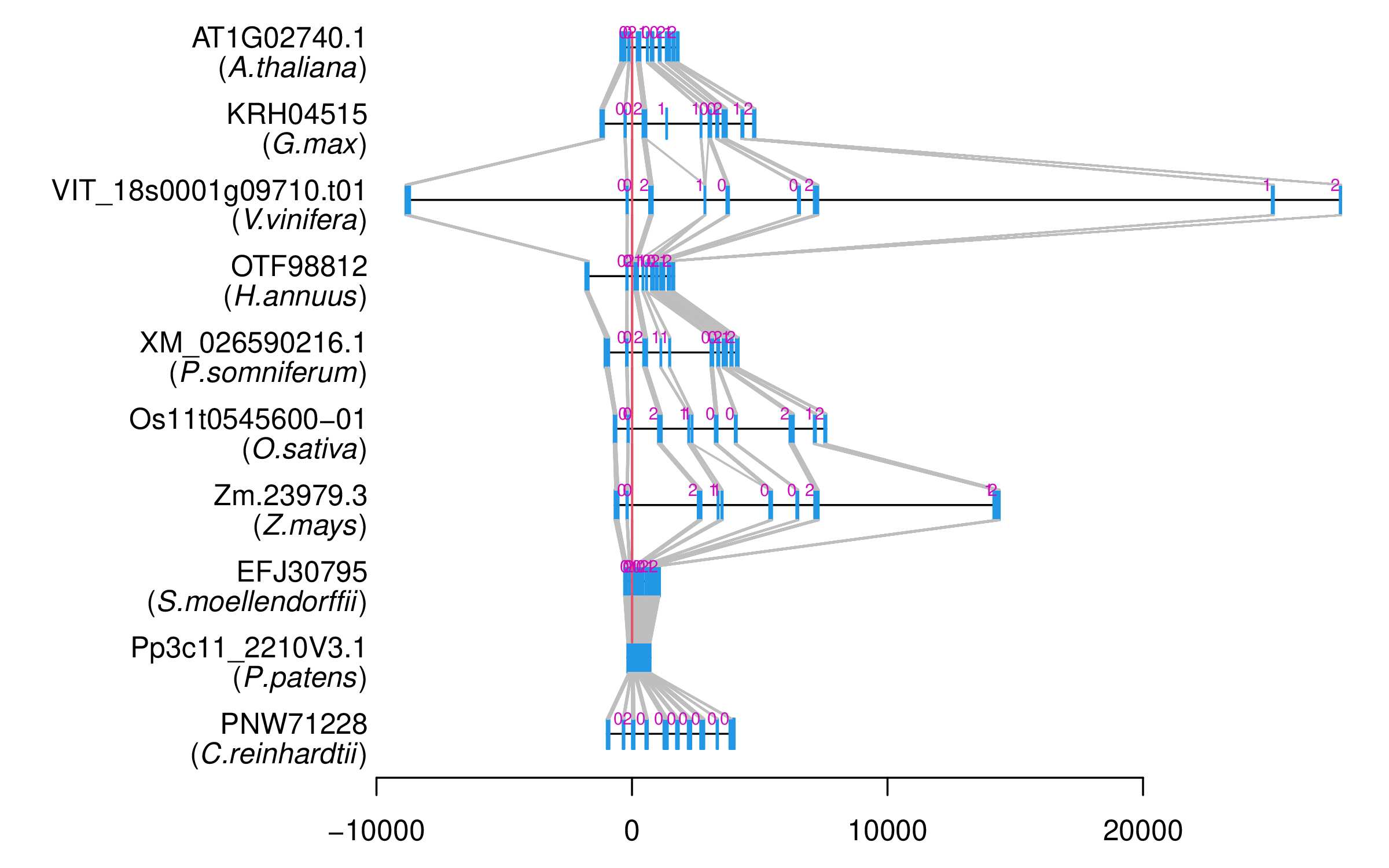 |
| Microexon DNA seq | GGTTGGAACAAAAA |
| Microexon Amino Acid seq | GWNKN |
| Microexon-tag DNA Seq | AAGGTCGAGCTTCGGAAAAAGGAGTGGAAATACTTTGTGCACTATCTTGGTTGGAACAAAAATTGGGATGAATGGGTTAGTGCGGATAGATTGTTGAAACATACCGAG |
| Microexon-tag Amino Acid Seq | KVELRKKEWKYFVHYLGWNKNWDEWVSADRLLKHTE |
| Microexon-tag spanning region | 17548317-17548623 |
| Microexon-tag prediction score | 0.9654 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | AT4G37280.1x |
| Reference Transcript ID | AT4G37280.1 |
| Gene ID | AT4G37280 |
| Gene Name | MRG1 |
| Transcript ID | AT4G37280.1 |
| Protein ID | AT4G37280.1 |
| Gene ID | AT4G37280 |
| Gene Name | MRG1 |
| Pfam domain motif | Tudor-knot |
| Motif E-value | 2.4e-11 |
| Motif start | 31 |
| Motif end | 80 |
| Protein seq | >AT4G37280.1 MGSSSKEETASDGDTASGGASPSNDGRLFSEGERVLAYHGPRVYGAKVQKVELRKKEWKYFVHYLGWNKNWDEWVSADRL LKHTEENLVKQKALDKKQGVEKGTKSGRSAQTKTRSSADTKADKDDTKTNAAKGKKRKHESGNEKDNVTAEKLMKIQIPA SLKKQLTDDWEYIAQKDKVVKLPRSPNVDEILSKYLEFKTKKDGMVTDSVAEILKGIRSYFDKALPVMLLYKKERRQYQE SIVDDTSPSTVYGAEHLLRLFVKLPDLFSYVNMEEETWSRMQQTLSDFLKFIQKNQSTFLLPSAYDSDKVSDGKGKGKDD * |
| CDS seq | >AT4G37280.1 ATGGGAAGCTCGTCGAAGGAAGAGACGGCGAGTGACGGAGATACGGCGAGCGGTGGCGCCTCACCGTCCAACGACGGCAG ACTCTTCTCGGAGGGCGAGCGTGTTCTCGCTTACCATGGTCCTCGCGTCTACGGAGCCAAGGTTCAAAAGGTCGAGCTTC GGAAAAAGGAGTGGAAATACTTTGTGCACTATCTTGGTTGGAACAAAAATTGGGATGAATGGGTTAGTGCGGATAGATTG TTGAAACATACCGAGGAGAATCTTGTCAAACAGAAAGCTCTAGACAAAAAGCAAGGAGTTGAAAAAGGTACCAAGTCTGG CCGTTCCGCTCAGACAAAAACTAGAAGCTCTGCAGACACAAAGGCTGATAAAGATGACACCAAGACCAATGCTGCAAAAG GGAAGAAACGAAAGCACGAGTCAGGCAATGAGAAGGATAATGTAACGGCGGAGAAGCTAATGAAGATCCAAATTCCCGCA TCACTTAAAAAGCAACTCACTGACGACTGGGAGTATATTGCGCAGAAGGACAAGGTTGTGAAACTTCCTCGTTCACCTAA CGTCGATGAGATATTGTCCAAGTACCTTGAATTCAAAACCAAGAAGGATGGAATGGTAACTGATTCGGTCGCTGAAATAT TGAAAGGCATAAGGAGTTATTTCGACAAGGCATTGCCAGTGATGCTCCTATATAAGAAAGAGAGGCGACAATACCAAGAA TCAATCGTGGATGATACTTCACCTTCAACTGTTTATGGCGCCGAGCATTTGCTCCGTCTCTTTGTCAAGTTGCCAGATCT TTTTTCATATGTAAACATGGAAGAAGAAACATGGAGTCGCATGCAACAAACATTATCAGACTTCCTCAAGTTTATTCAGA AGAATCAGAGCACTTTCTTGCTACCATCAGCATATGATTCTGATAAGGTTTCAGATGGTAAAGGCAAAGGAAAAGACGAC TGA |
| Microexon DNA seq | GGTTGGAACAAAAA |
| Microexon Amino Acid seq | GWNKN |
| Microexon-tag DNA Seq | AAGGTCGAGCTTCGGAAAAAGGAGTGGAAATACTTTGTGCACTATCTTGGTTGGAACAAAAATTGGGATGAATGGGTTAGTGCGGATAGATTGTTGAAACATACCGAG |
| Microexon-tag Amino Acid seq | KVELRKKEWKYFVHYLGWNKNWDEWVSADRLLKHTE |
| Transcript ID | AT4G37280.1 |
| Gene ID | At.21316 |
| Gene Name | MRG1 |
| Pfam domain motif | Tudor-knot |
| Motif E-value | 2.5e-11 |
| Motif start | 31 |
| Motif end | 80 |
| Protein seq | >AT4G37280.1 MGSSSKEETASDGDTASGGASPSNDGRLFSEGERVLAYHGPRVYGAKVQKVELRKKEWKYFVHYLGWNKNWDEWVSADRL LKHTEENLVKQKALDKKQGVEKGTKSGRSAQTKTRSSADTKADKDDTKTNAAKGKKRKHESGNEKDNVTAEKLMKIQIPA SLKKQLTDDWEYIAQKDKVVKLPRSPNVDEILSKYLEFKTKKDGMVTDSVAEILKGIRSYFDKALPVMLLYKKERRQYQE SIVDDTSPSTVYGAEHLLRLFVKLPDLFSYVNMEEETWSRMQQTLSDFLKFIQKNQSTFLLPSAYDSDKVSDGKGKGKDD * |
| CDS seq | >AT4G37280.1 ATGGGAAGCTCGTCGAAGGAAGAGACGGCGAGTGACGGAGATACGGCGAGCGGTGGCGCCTCACCGTCCAACGACGGCAG ACTCTTCTCGGAGGGCGAGCGTGTTCTCGCTTACCATGGTCCTCGCGTCTACGGAGCCAAGGTTCAAAAGGTCGAGCTTC GGAAAAAGGAGTGGAAATACTTTGTGCACTATCTTGGTTGGAACAAAAATTGGGATGAATGGGTTAGTGCGGATAGATTG TTGAAACATACCGAGGAGAATCTTGTCAAACAGAAAGCTCTAGACAAAAAGCAAGGAGTTGAAAAAGGTACCAAGTCTGG CCGTTCCGCTCAGACAAAAACTAGAAGCTCTGCAGACACAAAGGCTGATAAAGATGACACCAAGACCAATGCTGCAAAAG GGAAGAAACGAAAGCACGAGTCAGGCAATGAGAAGGATAATGTAACGGCGGAGAAGCTAATGAAGATCCAAATTCCCGCA TCACTTAAAAAGCAACTCACTGACGACTGGGAGTATATTGCGCAGAAGGACAAGGTTGTGAAACTTCCTCGTTCACCTAA CGTCGATGAGATATTGTCCAAGTACCTTGAATTCAAAACCAAGAAGGATGGAATGGTAACTGATTCGGTCGCTGAAATAT TGAAAGGCATAAGGAGTTATTTCGACAAGGCATTGCCAGTGATGCTCCTATATAAGAAAGAGAGGCGACAATACCAAGAA TCAATCGTGGATGATACTTCACCTTCAACTGTTTATGGCGCCGAGCATTTGCTCCGTCTCTTTGTCAAGTTGCCAGATCT TTTTTCATATGTAAACATGGAAGAAGAAACATGGAGTCGCATGCAACAAACATTATCAGACTTCCTCAAGTTTATTCAGA AGAATCAGAGCACTTTCTTGCTACCATCAGCATATGATTCTGATAAGGTTTCAGATGGTAAAGGCAAAGGAAAAGACGAC TGA |