
| Microexon ID | At_1:601547-601560:- |
| Species | Arabidopsis thaliana | Coordinates | 1:601547..601560 |
| Microexon Cluster ID | MEP35 |
| Size | 14 |
| Phase | 0 |
| Pfam Domain Motif | Tudor-knot |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,14,46 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | AARGCTGARHTYCRRAAGAAKGARTGGARATAYTTYGTYCATTAYCTTGGTTGGAAYAAAAATTGGGATGAATGGGTWGGYRYKGAYCGYYTGWTGAARYWYACTGAR |
| Logo of Microexon-tag DNA Seq | 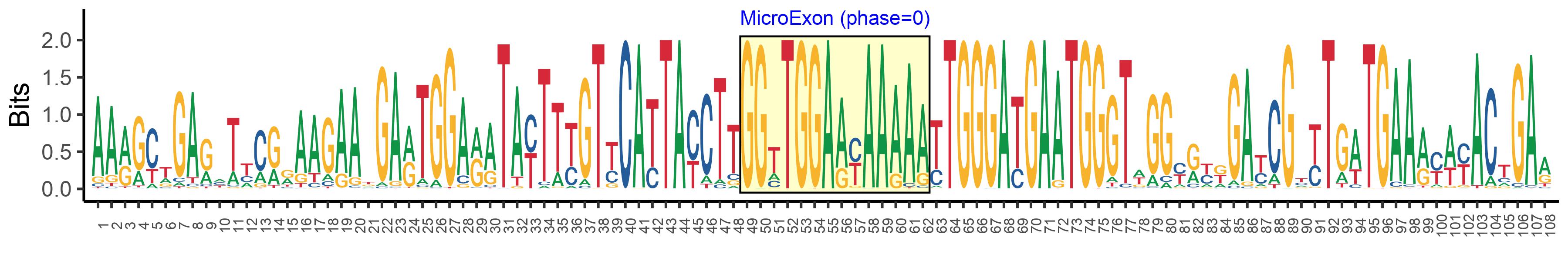 |
| Alignment of exons | 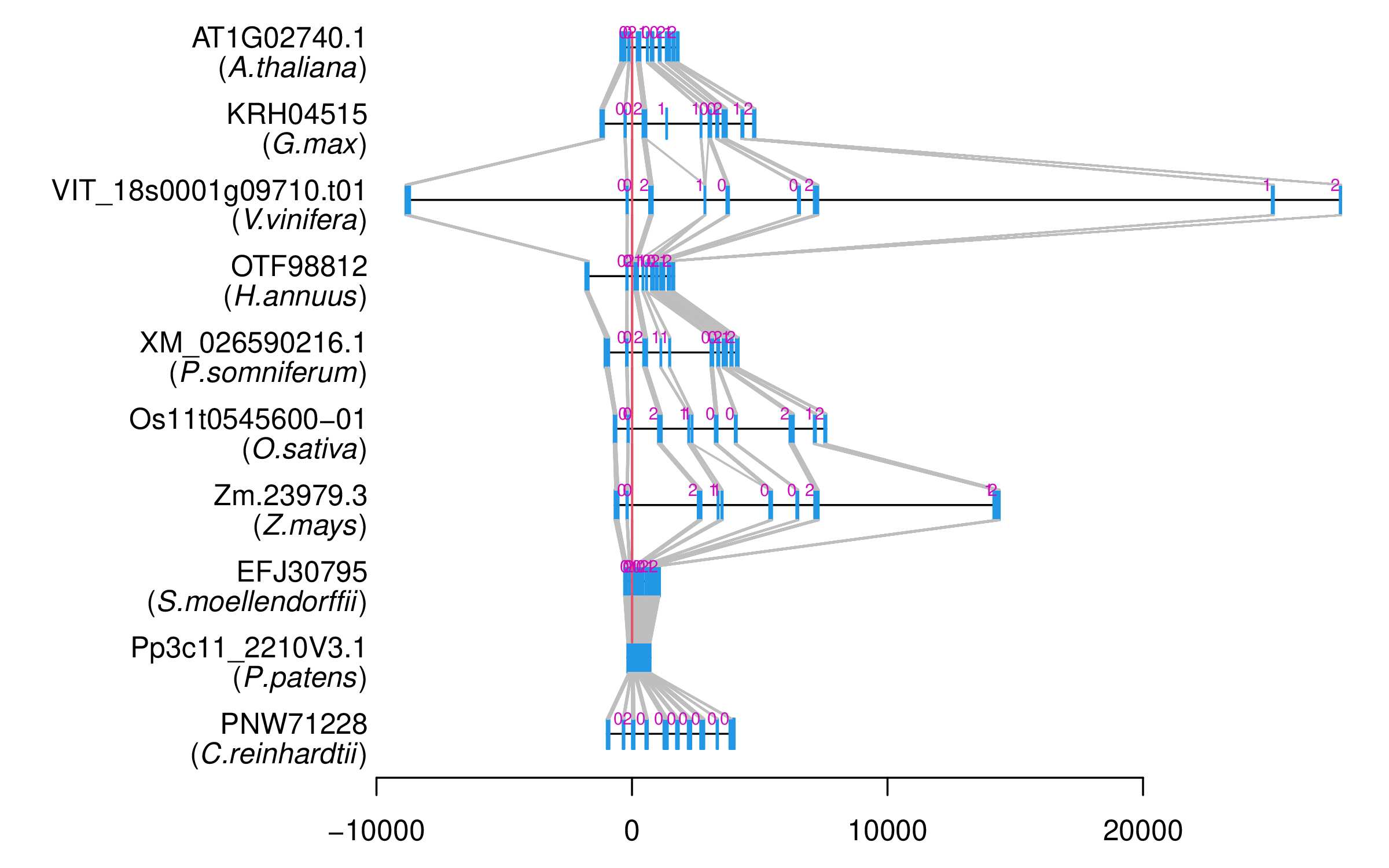 |
| Microexon DNA seq | GGTTGGAACAAAAG |
| Microexon Amino Acid seq | GWNKS |
| Microexon-tag DNA Seq | AAAGTTGAATTTAAAGACAATGAATGGAAATATTTTGTGCATTACATTGGTTGGAACAAAAGTTGGGACGAATGGATAAGGCTGGACTGTCTGTTGAAACATTCAGAT |
| Microexon-tag Amino Acid Seq | KVEFKDNEWKYFVHYIGWNKSWDEWIRLDCLLKHSD |
| Microexon-tag spanning region | 601327-601700 |
| Microexon-tag prediction score | 0.9228 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | AT1G02740.1x |
| Reference Transcript ID | AT1G02740.1 |
| Gene ID | AT1G02740 |
| Gene Name | MRG2 |
| Transcript ID | AT1G02740.1 |
| Protein ID | AT1G02740.1 |
| Gene ID | AT1G02740 |
| Gene Name | MRG2 |
| Pfam domain motif | Tudor-knot |
| Motif E-value | 1.8e-11 |
| Motif start | 53 |
| Motif end | 101 |
| Protein seq | >AT1G02740.1 MGSPNAAAETDLTTDDFIGDTRRDSGSDTETNTDCDGEDLPLLLLPAPPGHFEEGERVLAKHSDCFYEAKVLKVEFKDNE WKYFVHYIGWNKSWDEWIRLDCLLKHSDENIEKQKEQGLKQQGIKSAMAWKVSKMKPRSPNVARGRKRKQDSVDTEKNVL PSDNLLSFNIPPALRKQLLDDFEFVTQMQKLVQLPRSPNVDGILKKYIDSQMKKHGRVTDSLEEILKGLRCYFDKALPVM LLYNNERKQYEESVSGGVSPSTVYGAEHLLRLFVKLPELLVHVNMAEETLKELQDNFVDILRFLRKNQSVLFVSTYKAVE EMEKKEG* |
| CDS seq | >AT1G02740.1 ATGGGAAGCCCTAACGCCGCCGCCGAGACGGATCTAACAACCGATGATTTCATCGGAGACACACGACGGGACTCTGGTTC TGATACAGAGACCAACACCGATTGTGACGGAGAAGATTTGCCTCTTCTTCTGCTACCTGCTCCTCCCGGCCACTTCGAAG AAGGAGAGAGAGTTTTAGCCAAACACAGTGATTGCTTCTACGAAGCCAAGGTTCTTAAAGTTGAATTTAAAGACAATGAA TGGAAATATTTTGTGCATTACATTGGTTGGAACAAAAGTTGGGACGAATGGATAAGGCTGGACTGTCTGTTGAAACATTC AGATGAGAATATTGAGAAACAGAAAGAACAGGGTTTGAAACAGCAAGGGATTAAGAGTGCTATGGCTTGGAAAGTGTCAA AGATGAAACCTAGAAGCCCTAATGTTGCTAGAGGAAGAAAGCGGAAGCAAGATTCTGTTGATACGGAGAAGAATGTGCTT CCCTCTGACAACCTTTTAAGTTTCAATATTCCACCAGCATTAAGGAAGCAACTTCTTGACGATTTTGAATTTGTTACTCA GATGCAGAAGCTTGTGCAACTTCCGCGTTCTCCAAATGTGGATGGCATCTTGAAGAAGTACATTGACAGCCAAATGAAGA AACATGGCAGGGTAACAGATTCATTAGAGGAAATTCTGAAAGGCTTGCGTTGCTACTTTGACAAAGCTTTACCGGTGATG TTGCTTTACAACAATGAACGGAAGCAGTACGAGGAATCCGTATCAGGGGGTGTTTCTCCTTCAACTGTGTACGGAGCAGA GCATTTGTTGCGACTCTTTGTGAAATTACCGGAGTTGCTAGTACATGTGAATATGGCAGAAGAGACATTGAAGGAATTGC AGGACAACTTTGTTGATATTCTCAGGTTCTTGAGGAAGAATCAGAGTGTGCTTTTTGTATCGACATACAAAGCAGTGGAA GAAATGGAGAAGAAAGAAGGTTAG |
| Microexon DNA seq | GGTTGGAACAAAAG |
| Microexon Amino Acid seq | GWNKS |
| Microexon-tag DNA Seq | AAAGTTGAATTTAAAGACAATGAATGGAAATATTTTGTGCATTACATTGGTTGGAACAAAAGTTGGGACGAATGGATAAGGCTGGACTGTCTGTTGAAACATTCAGAT |
| Microexon-tag Amino Acid seq | KVEFKDNEWKYFVHYIGWNKSWDEWIRLDCLLKHSD |
| Transcript ID | AT1G02740.1 |
| Gene ID | At.188 |
| Gene Name | MRG2 |
| Pfam domain motif | Tudor-knot |
| Motif E-value | 1.8e-11 |
| Motif start | 53 |
| Motif end | 101 |
| Protein seq | >AT1G02740.1 MGSPNAAAETDLTTDDFIGDTRRDSGSDTETNTDCDGEDLPLLLLPAPPGHFEEGERVLAKHSDCFYEAKVLKVEFKDNE WKYFVHYIGWNKSWDEWIRLDCLLKHSDENIEKQKEQGLKQQGIKSAMAWKVSKMKPRSPNVARGRKRKQDSVDTEKNVL PSDNLLSFNIPPALRKQLLDDFEFVTQMQKLVQLPRSPNVDGILKKYIDSQMKKHGRVTDSLEEILKGLRCYFDKALPVM LLYNNERKQYEESVSGGVSPSTVYGAEHLLRLFVKLPELLVHVNMAEETLKELQDNFVDILRFLRKNQSVLFVSTYKAVE EMEKKEG* |
| CDS seq | >AT1G02740.1 ATGGGAAGCCCTAACGCCGCCGCCGAGACGGATCTAACAACCGATGATTTCATCGGAGACACACGACGGGACTCTGGTTC TGATACAGAGACCAACACCGATTGTGACGGAGAAGATTTGCCTCTTCTTCTGCTACCTGCTCCTCCCGGCCACTTCGAAG AAGGAGAGAGAGTTTTAGCCAAACACAGTGATTGCTTCTACGAAGCCAAGGTTCTTAAAGTTGAATTTAAAGACAATGAA TGGAAATATTTTGTGCATTACATTGGTTGGAACAAAAGTTGGGACGAATGGATAAGGCTGGACTGTCTGTTGAAACATTC AGATGAGAATATTGAGAAACAGAAAGAACAGGGTTTGAAACAGCAAGGGATTAAGAGTGCTATGGCTTGGAAAGTGTCAA AGATGAAACCTAGAAGCCCTAATGTTGCTAGAGGAAGAAAGCGGAAGCAAGATTCTGTTGATACGGAGAAGAATGTGCTT CCCTCTGACAACCTTTTAAGTTTCAATATTCCACCAGCATTAAGGAAGCAACTTCTTGACGATTTTGAATTTGTTACTCA GATGCAGAAGCTTGTGCAACTTCCGCGTTCTCCAAATGTGGATGGCATCTTGAAGAAGTACATTGACAGCCAAATGAAGA AACATGGCAGGGTAACAGATTCATTAGAGGAAATTCTGAAAGGCTTGCGTTGCTACTTTGACAAAGCTTTACCGGTGATG TTGCTTTACAACAATGAACGGAAGCAGTACGAGGAATCCGTATCAGGGGGTGTTTCTCCTTCAACTGTGTACGGAGCAGA GCATTTGTTGCGACTCTTTGTGAAATTACCGGAGTTGCTAGTACATGTGAATATGGCAGAAGAGACATTGAAGGAATTGC AGGACAACTTTGTTGATATTCTCAGGTTCTTGAGGAAGAATCAGAGTGTGCTTTTTGTATCGACATACAAAGCAGTGGAA GAAATGGAGAAGAAAGAAGGTTAG |