
| Microexon ID | Zm_10:15574035-15574048:+ |
| Species | Zea mays | Coordinates | 10:15574035..15574048 |
| Microexon Cluster ID | MEP34 |
| Size | 14 |
| Phase | 0 |
| Pfam Domain Motif | DUF1325 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,14,46 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GGAACMACWGCACTTTAYAARGCAACTGTWGTTARTACYCMTCGMAAGAGGAARWCTGAYGAKTATTTGYTGGARTTYGATGAYGAYGARGAAGAYGGAKCTTTGCCK |
| Logo of Microexon-tag DNA Seq | 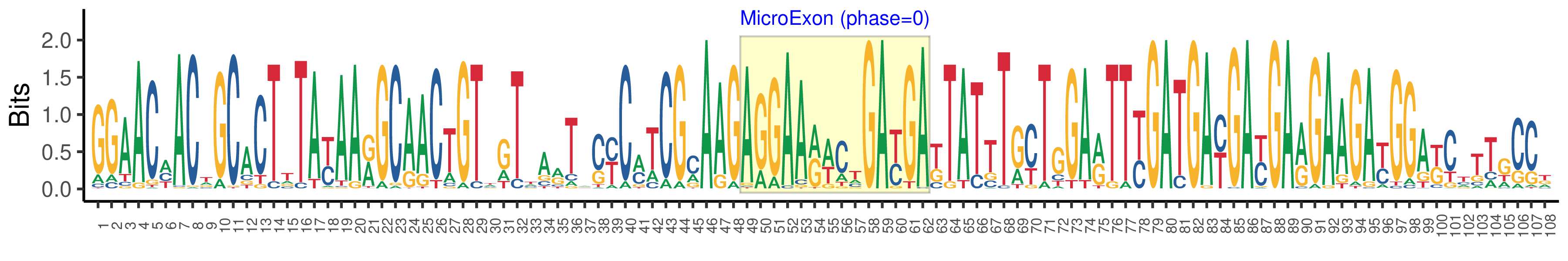 |
| Alignment of exons | 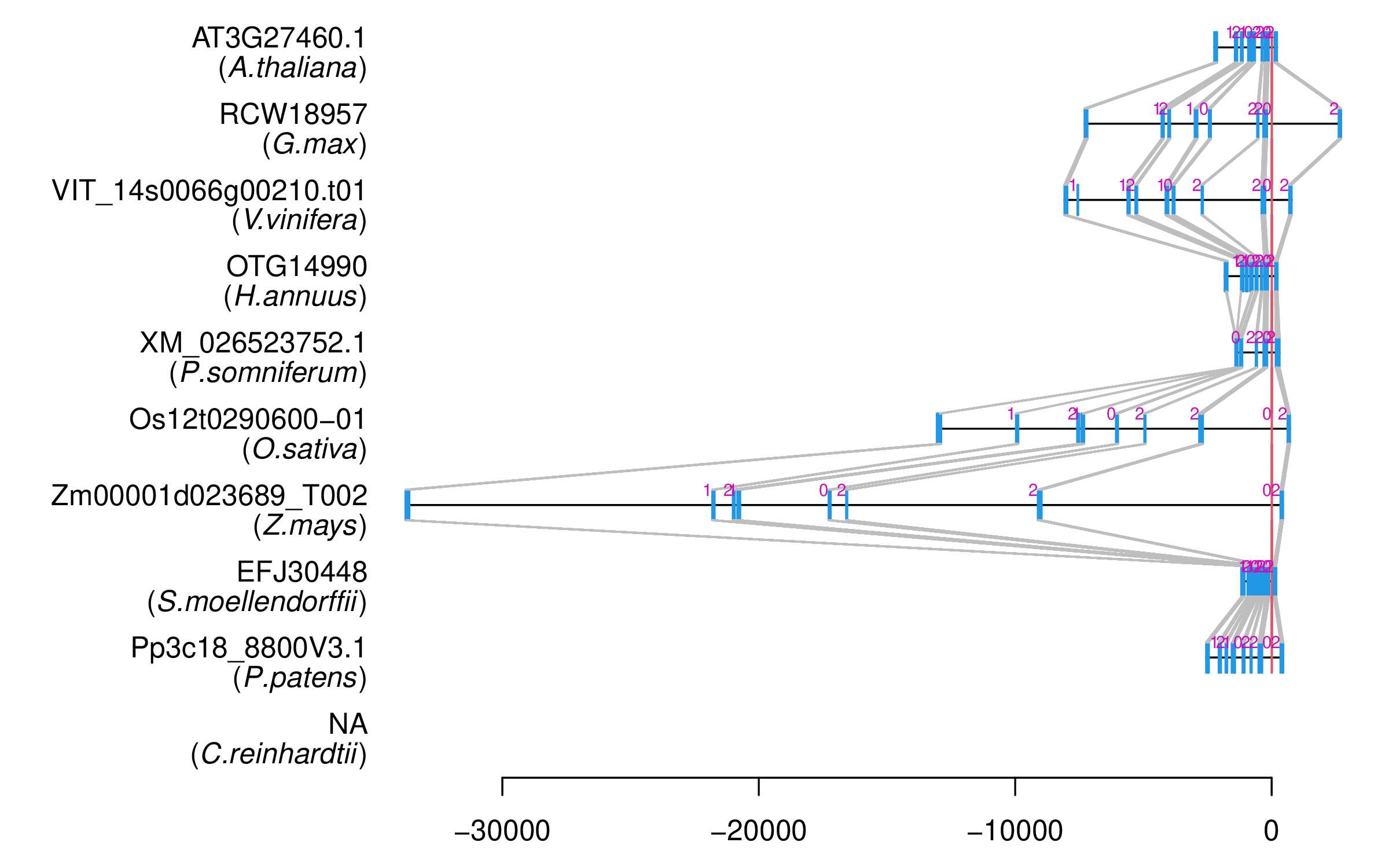 |
Zm_10:15574035-15574048:+ does not have available information here.
| Transcript ID | Zm00001d023689_T001 |
| Protein ID | NA |
| Gene ID | Zm00001d023689 |
| Gene Name | NA |
| Pfam domain motif | NA |
| Motif E-value | NA |
| Motif start | NA |
| Motif end | NA |
| Protein seq | |
| CDS seq |
| Microexon DNA seq | AGGAAGTCTGATGA |
| Microexon Amino Acid seq | RKSDD |
| Microexon-tag DNA Seq | CCTGGCACAACAGCATTATACAGAGCTACCGTAGCTTCTCATCGTAAGAGGAAGTCTGATGATTATATTCTGGAGTTCGATGACGATGAGGAGGACGGTGCTCTGCCG |
| Microexon-tag Amino Acid seq | PGTTALYRATVASHRKRKSDDYILEFDDDEEDGALP |
| Transcript ID | Zm00001d023689_T002 |
| Gene ID | Zm.6255 |
| Gene Name | NA |
| Pfam domain motif | DUF1325 |
| Motif E-value | 1.9e-34 |
| Motif start | 142 |
| Motif end | 273 |
| Protein seq | >Zm00001d023689_T002 MSSSVGGGSAGGGGVDMVSLLEKAKDLDQLKKEQDEIVADINKIHKKLLASPEMVDKPVDTILLKLRGLYARAKELSENE ISASNALIGLLDGLLQSGASTAQKKKIEVGEQKKKRIKSDTDSARFSAASMRNQLDQAASLKGEQVAARVKSDDEKDEWF VVKVIHFDKETKEYEVLDEEPGDDEESTQKKYKLPMSYIIPFPKKGDPSSAPDFGQGRQVLAVYPGTTALYRATVASHRK RKSDDYILEFDDDEEDGALPQRAVPFYRVVALPEGHRQ* |
| CDS seq | >Zm00001d023689_T002 ATGTCGTCGTCGGTTGGGGGCGGCAGCGCGGGCGGCGGCGGAGTGGACATGGTCAGCCTGCTGGAGAAGGCCAAGGATTT GGACCAGCTAAAGAAGGAACAGGACGAAATCGTTGCCGATATCAACAAGATTCACAAGAAGCTCCTCGCATCCCCAGAGA TGGTTGATAAACCAGTTGACACCATTTTGCTCAAGCTTCGTGGTTTATATGCACGTGCAAAGGAGCTCTCTGAGAATGAA ATCAGTGCCTCAAATGCTTTGATTGGATTATTAGATGGTTTGTTGCAATCTGGAGCTTCAACTGCACAAAAGAAAAAGAT AGAAGTTGGTGAGCAGAAAAAGAAGAGGATCAAGTCTGATACAGATTCTGCACGATTTTCTGCTGCATCTATGAGGAATC AACTTGACCAAGCTGCAAGCCTTAAAGGAGAGCAGGTTGCTGCTAGGGTAAAATCAGATGACGAAAAAGATGAGTGGTTT GTCGTGAAGGTCATACATTTCGATAAGGAAACAAAAGAGTATGAAGTACTTGATGAAGAACCAGGTGATGACGAAGAGAG CACTCAAAAGAAGTACAAGTTGCCTATGTCTTACATTATTCCATTTCCGAAGAAAGGGGATCCTTCGAGTGCCCCGGATT TTGGGCAGGGTCGACAAGTTTTAGCTGTTTATCCTGGCACAACAGCATTATACAGAGCTACCGTAGCTTCTCATCGTAAG AGGAAGTCTGATGATTATATTCTGGAGTTCGATGACGATGAGGAGGACGGTGCTCTGCCGCAGAGAGCCGTGCCGTTCTA CAGGGTGGTTGCCCTGCCGGAGGGCCACAGGCAGTAG |