
| Microexon ID | Vv_14:26776303-26776316:+ |
| Species | Vistis vinifera | Coordinates | 14:26776303..26776316 |
| Microexon Cluster ID | MEP34 |
| Size | 14 |
| Phase | 0 |
| Pfam Domain Motif | DUF1325 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,14,46 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GGAACMACWGCACTTTAYAARGCAACTGTWGTTARTACYCMTCGMAAGAGGAARWCTGAYGAKTATTTGYTGGARTTYGATGAYGAYGARGAAGAYGGAKCTTTGCCK |
| Logo of Microexon-tag DNA Seq | 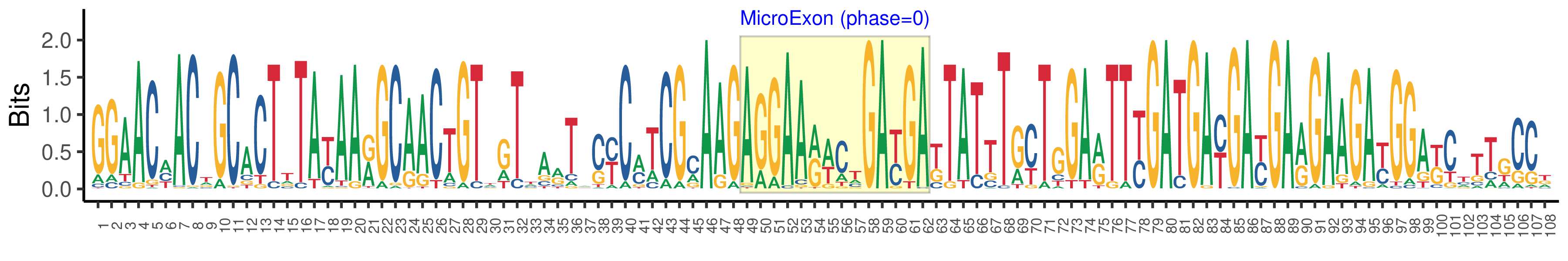 |
| Alignment of exons | 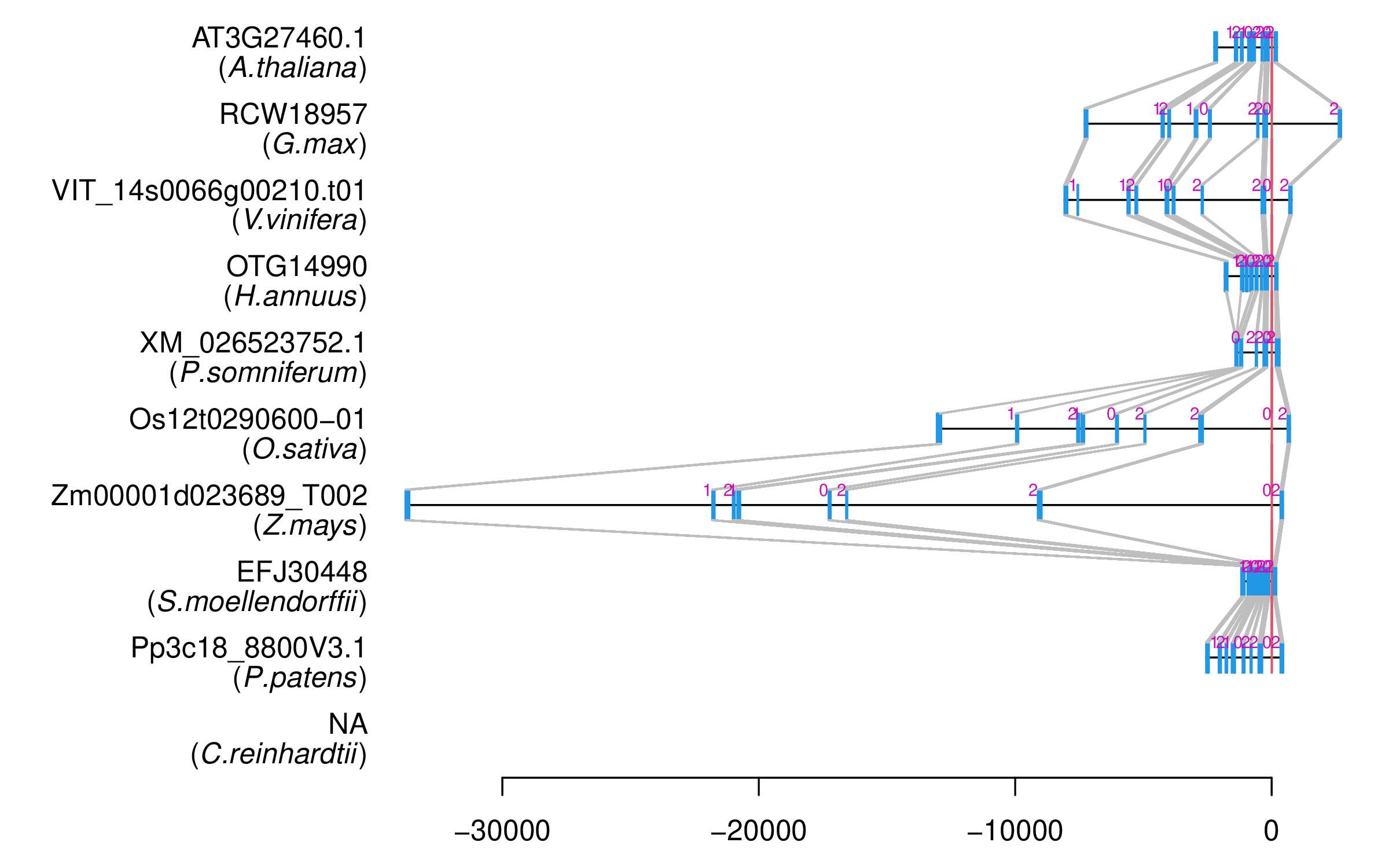 |
| Microexon DNA seq | AGGAAGACCGATGA |
| Microexon Amino Acid seq | RKTDD |
| Microexon-tag DNA Seq | GGGACAACTGCACTTTATAAAGCAACTGTTGTCAATCCCCATCGTAAGAGGAAGACCGATGATTATGTACTGGAATTCGATGACGACGAGGAAGATGGATCTTTACCC |
| Microexon-tag Amino Acid Seq | GTTALYKATVVNPHRKRKTDDYVLEFDDDEEDGSLP |
| Microexon-tag spanning region | 26776011-26777030 |
| Microexon-tag prediction score | 0.9716 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | VIT_14s0066g00210.t01x |
| Reference Transcript ID | VIT_14s0066g00210.t01 |
| Gene ID | VIT_14s0066g00210 |
| Gene Name | NA |
| Transcript ID | VIT_14s0066g00210.t01 |
| Protein ID | VIT_14s0066g00210.t01 |
| Gene ID | VIT_14s0066g00210 |
| Gene Name | NA |
| Pfam domain motif | DUF1325 |
| Motif E-value | 1.7e-33 |
| Motif start | 142 |
| Motif end | 276 |
| Protein seq | >VIT_14s0066g00210.t01 MSSPDITGILETSKDLDRLRREQDEILQEINKLHKKLQSGREKSGIKMAGAPEMVEKPTDSSLSKLRALYYQARDLSENE VSVSNQLLAQLDALLPSAPAPGQQRRRIDGSEQKKKRMKTDSDISRPSPSIRNQLEVAASLKGEQVAARVTPDDADKDEW FVVKVIHFDRDAREFEVLDEEPGDDEESSGQRKYKLPMSHIIPFPKRNDTNGVPDFPNGRQVLAVYPGTTALYKATVVNP HRKRKTDDYVLEFDDDEEDGSLPQRVVPFHKVVALPEGHRQ* |
| CDS seq | >VIT_14s0066g00210.t01 ATGTCGTCGCCAGACATTACCGGAATCCTCGAAACCTCGAAGGATCTGGATCGATTGAGGAGAGAGCAAGACGAGATACT CCAGGAGATAAACAAGTTGCATAAAAAGCTTCAATCTGGAAGAGAAAAGAGTGGAATTAAGATGGCTGGAGCTCCTGAGA TGGTTGAGAAGCCTACAGATAGTTCACTATCAAAGCTTAGGGCTTTATATTATCAGGCCAGGGATCTTTCAGAGAATGAA GTGAGTGTTTCCAATCAGTTGTTAGCTCAGCTGGATGCTCTGCTGCCTTCTGCACCTGCTCCTGGGCAACAACGAAGAAG GATAGATGGTAGCGAGCAGAAAAAGAAACGGATGAAGACTGATTCAGATATCTCTCGGCCTTCTCCATCCATTCGAAACC AACTTGAGGTTGCTGCTAGTCTAAAGGGTGAACAGGTAGCTGCTCGGGTCACTCCAGATGATGCTGACAAGGATGAATGG TTTGTTGTAAAAGTAATACATTTTGATCGGGACGCAAGAGAATTTGAAGTACTTGATGAGGAACCTGGTGATGATGAAGA GAGTAGTGGCCAAAGGAAGTATAAGCTGCCTATGTCGCATATTATACCTTTTCCCAAGCGAAATGACACTAACGGTGTCC CAGATTTCCCTAATGGAAGGCAAGTTTTGGCAGTCTATCCAGGGACAACTGCACTTTATAAAGCAACTGTTGTCAATCCC CATCGTAAGAGGAAGACCGATGATTATGTACTGGAATTCGATGACGACGAGGAAGATGGATCTTTACCCCAAAGGGTAGT GCCCTTCCACAAGGTGGTTGCCCTGCCGGAGGGACATCGCCAGTGA |
| Microexon DNA seq | AGGAAGACCGATGA |
| Microexon Amino Acid seq | RKTDD |
| Microexon-tag DNA Seq | GGGACAACTGCACTTTATAAAGCAACTGTTGTCAATCCCCATCGTAAGAGGAAGACCGATGATTATGTACTGGAATTCGATGACGACGAGGAAGATGGATCTTTACCC |
| Microexon-tag Amino Acid seq | GTTALYKATVVNPHRKRKTDDYVLEFDDDEEDGSLP |
| Transcript ID | Vv.10169.1 |
| Gene ID | Vv.10169 |
| Gene Name | NA |
| Pfam domain motif | DUF1325 |
| Motif E-value | 1.5e-33 |
| Motif start | 131 |
| Motif end | 265 |
| Protein seq | >Vv.10169.1 MSSPDITGILETSKDLDRLRREQDEILQEINKLHKKLQSAPEMVEKPTDSSLSKLRALYYQARDLSENEVSVSNQLLAQL DALLPSAPAPGQQRRRIDGSEQKKKRMKTDSDISRPSPSIRNQLEVAASLKGEQVAARVTPDDADKDEWFVVKVIHFDRD AREFEVLDEEPGDDEESSGQRKYKLPMSHIIPFPKRNDTNGVPDFPNGRQVLAVYPGTTALYKATVVNPHRKRKTDDYVL EFDDDEEDGSLPQRVVPFHKVVALPEGHRQ* |
| CDS seq | >Vv.10169.1 ATGTCGTCGCCAGACATTACCGGAATCCTCGAAACCTCGAAGGATCTGGATCGATTGAGGAGAGAGCAAGACGAGATACT CCAGGAGATAAACAAGTTGCATAAAAAGCTTCAATCTGCTCCTGAGATGGTTGAGAAGCCTACAGATAGTTCACTATCAA AGCTTAGGGCTTTATATTATCAGGCCAGGGATCTTTCAGAGAATGAAGTGAGTGTTTCCAATCAGTTGTTAGCTCAGCTG GATGCTCTGCTGCCTTCTGCACCTGCTCCTGGGCAACAACGAAGAAGGATAGATGGTAGCGAGCAGAAAAAGAAACGGAT GAAGACTGATTCAGATATCTCTCGGCCTTCTCCATCCATTCGAAACCAACTTGAGGTTGCTGCTAGTCTAAAGGGTGAAC AGGTAGCTGCTCGGGTCACTCCAGATGATGCTGACAAGGATGAATGGTTTGTTGTAAAAGTAATACATTTTGATCGGGAC GCAAGAGAATTTGAAGTACTTGATGAGGAACCTGGTGATGATGAAGAGAGTAGTGGCCAAAGGAAGTATAAGCTGCCTAT GTCGCATATTATACCTTTTCCCAAGCGAAATGACACTAACGGTGTCCCAGATTTCCCTAATGGAAGGCAAGTTTTGGCAG TCTATCCAGGGACAACTGCACTTTATAAAGCAACTGTTGTCAATCCCCATCGTAAGAGGAAGACCGATGATTATGTACTG GAATTCGATGACGACGAGGAAGATGGATCTTTACCCCAAAGGGTAGTGCCCTTCCACAAGGTGGTTGCCCTGCCGGAGGG ACATCGCCAGTGA |