
| Microexon ID | Sm_GL377575:1682396-1682409:+ |
| Species | Selaginella moellendorffii | Coordinates | GL377575:1682396..1682409 |
| Microexon Cluster ID | MEP34 |
| Size | 14 |
| Phase | 0 |
| Pfam Domain Motif | DUF1325 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,14,46 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GGAACMACWGCACTTTAYAARGCAACTGTWGTTARTACYCMTCGMAAGAGGAARWCTGAYGAKTATTTGYTGGARTTYGATGAYGAYGARGAAGAYGGAKCTTTGCCK |
| Logo of Microexon-tag DNA Seq | 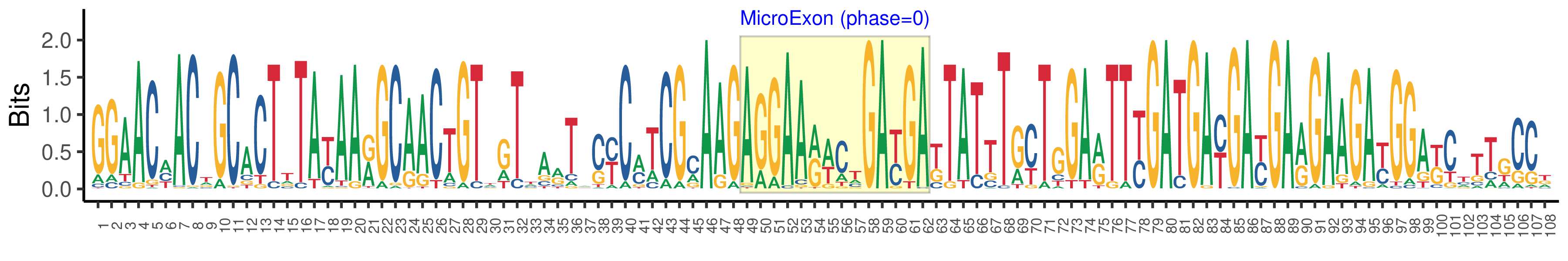 |
| Alignment of exons | 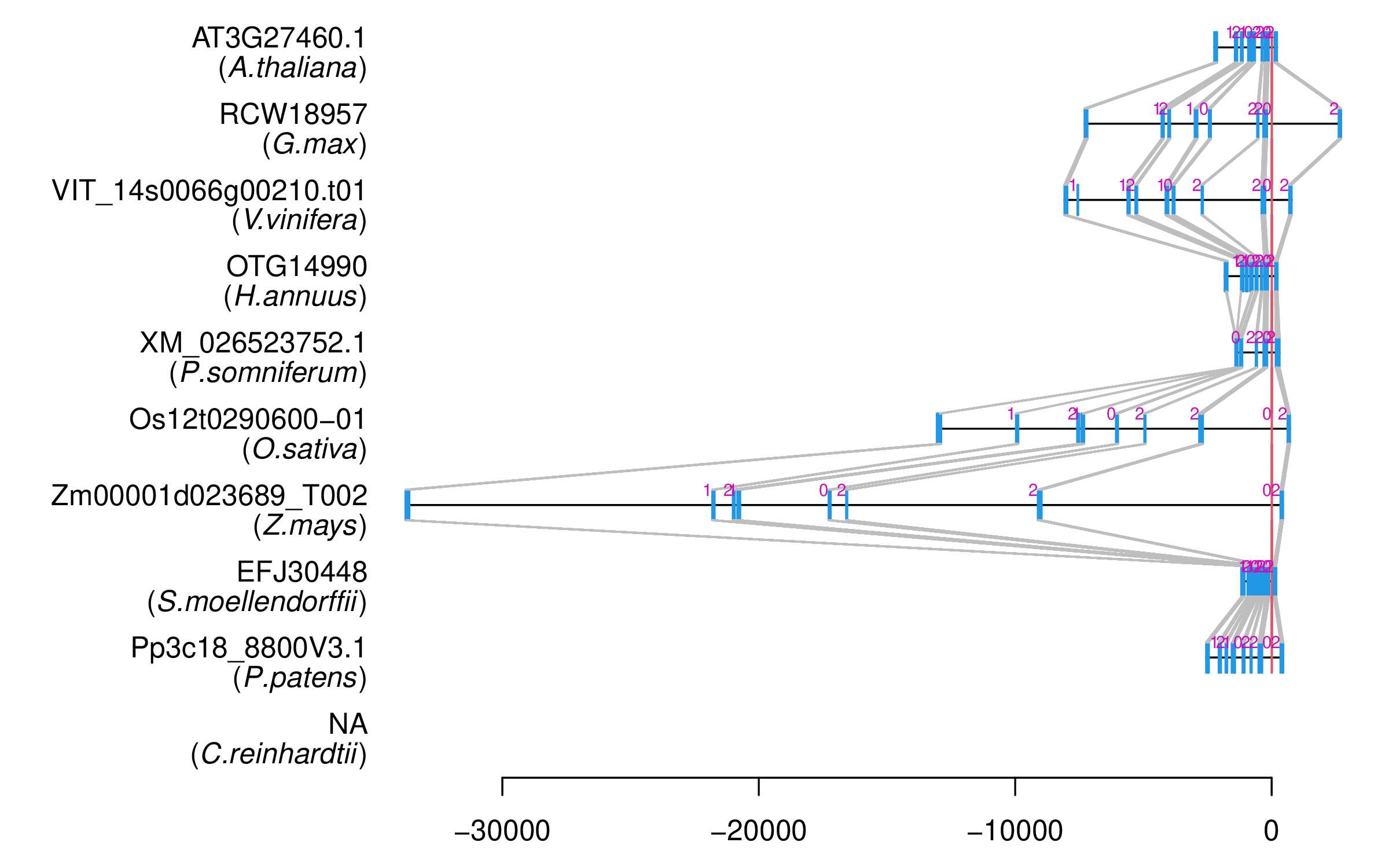 |
| Microexon DNA seq | AAAAAAACGGACGA |
| Microexon Amino Acid seq | KKTDD |
| Microexon-tag DNA Seq | GGCACCACCGCACTGTACAAAGCTACTGTCGTCGGTCCTCATAGAAAAAAAAAAACGGACGACTATATTCTAGAATTCGATGACGACGACGAGGATGGATTTCTACCA |
| Microexon-tag Amino Acid Seq | GTTALYKATVVGPHRKKKTDDYILEFDDDDEDGFLP |
| Microexon-tag spanning region | 1682263-1682520 |
| Microexon-tag prediction score | 0.9145 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | EFJ30448x |
| Reference Transcript ID | EFJ30448 |
| Gene ID | SELMODRAFT_89941 |
| Gene Name | NA |
| Transcript ID | EFJ30448 |
| Protein ID | EFJ30448 |
| Gene ID | SELMODRAFT_89941 |
| Gene Name | NA |
| Pfam domain motif | DUF1325 |
| Motif E-value | 2.1e-32 |
| Motif start | 138 |
| Motif end | 271 |
| Protein seq | >EFJ30448 MASNALSAASVEAASQGLKELDRLRREQDGLIKKINKIHAKLAETQPEQAEKFEDKFWSKLRDLYSQAKALADAEESASN ACLSNLDALSIPSQAAAAHRRKTETGDQKRKRAKADADSSRLNPNVSRSADSIGKAVGDQVAARITPEDSEKSEWIVVKV TRFDRETNKYEVIDEEPGDEEENGGTGGPRKYKLSPSAIIQFPKPATALDFPTGSQVLAVYPGTTALYKATVVGPHRKKK TDDYILEFDDDDEDGFLPKRTVPFYHVVTLPEGHRQ* |
| CDS seq | >EFJ30448 ATGGCGAGCAATGCATTGAGCGCGGCGAGCGTGGAGGCCGCGTCGCAAGGGCTCAAGGAGCTGGATCGATTGAGGAGAGA ACAAGATGGTCTGATCAAGAAGATCAATAAGATCCACGCCAAGCTCGCAGAGACCCAACCAGAGCAAGCCGAGAAGTTCG AAGATAAGTTCTGGAGCAAGCTACGGGATTTGTATAGTCAAGCTAAAGCACTTGCGGACGCCGAAGAAAGTGCGTCAAAT GCTTGTCTTAGTAATCTGGACGCTCTCTCGATACCTTCACAAGCTGCTGCTGCTCATCGACGTAAAACTGAGACTGGCGA TCAAAAGCGGAAGAGAGCCAAGGCCGATGCAGACTCTTCCAGGCTGAATCCGAACGTTTCTCGGTCGGCTGATTCAATTG GTAAAGCTGTTGGAGACCAGGTCGCCGCGCGGATTACTCCTGAAGATTCAGAGAAGAGCGAGTGGATTGTCGTCAAGGTG ACCCGCTTTGATCGCGAAACAAACAAATATGAAGTGATAGACGAAGAGCCTGGGGACGAAGAAGAAAATGGTGGAACCGG TGGACCGAGGAAGTACAAGTTATCTCCCTCCGCAATCATCCAATTCCCAAAACCAGCGACAGCGCTGGATTTTCCAACGG GATCTCAAGTTCTTGCGGTGTATCCAGGCACCACCGCACTGTACAAAGCTACTGTCGTCGGTCCTCATAGAAAAAAAAAA ACGGACGACTATATTCTAGAATTCGATGACGACGACGAGGATGGATTTCTACCAAAGCGAACAGTGCCATTTTACCACGT TGTCACTCTTCCGGAGGGGCACCGCCAGTAA |
| Microexon DNA seq | AAAAAAACGGACGA |
| Microexon Amino Acid seq | KKTDD |
| Microexon-tag DNA Seq | GGCACCACCGCACTGTACAAAGCTACTGTCGTCGGTCCTCATAGAAAAAAAAAAACGGACGACTATATTCTAGAATTCGATGACGACGACGAGGATGGATTTCTACCA |
| Microexon-tag Amino Acid seq | GTTALYKATVVGPHRKKKTDDYILEFDDDDEDGFLP |
| Transcript ID | Sm.7835.1 |
| Gene ID | Sm.7835 |
| Gene Name | NA |
| Pfam domain motif | DUF1325 |
| Motif E-value | 2.1e-32 |
| Motif start | 138 |
| Motif end | 271 |
| Protein seq | >Sm.7835.1 MASNALSAASVEAASQGLKELDRLRREQDGLIKKINKIHAKLAETQPEQAEKFEDKFWSKLRDLYSQAKALADAEESASN ACLSNLDALSIPSQAAAAHRRKTETGDQKRKRAKADADSSRLNPNVSRSADSIGKAVGDQVAARITPEDSEKSEWIVVKV TRFDRETNKYEVIDEEPGDEEENGGTGGPRKYKLSPSAIIQFPKPATALDFPTGSQVLAVYPGTTALYKATVVGPHRKKK TDDYILEFDDDDEDGFLPKRTVPFYHVVTLPEGHRQ* |
| CDS seq | >Sm.7835.1 ATGGCGAGCAATGCATTGAGCGCGGCGAGCGTGGAGGCCGCGTCGCAAGGGCTCAAGGAGCTGGATCGATTGAGGAGAGA ACAAGATGGTCTGATCAAGAAGATCAATAAGATCCACGCCAAGCTCGCAGAGACCCAACCAGAGCAAGCCGAGAAGTTCG AAGATAAGTTCTGGAGCAAGCTACGGGATTTGTATAGTCAAGCTAAAGCACTTGCGGACGCCGAAGAAAGTGCGTCAAAT GCTTGTCTTAGTAATCTGGACGCTCTCTCGATACCTTCACAAGCTGCTGCTGCTCATCGACGTAAAACTGAGACTGGCGA TCAAAAGCGGAAGAGAGCCAAGGCCGATGCAGACTCTTCCAGGCTGAATCCGAACGTTTCTCGGTCGGCTGATTCAATTG GTAAAGCTGTTGGAGACCAGGTCGCCGCGCGGATTACTCCTGAAGATTCAGAGAAGAGCGAGTGGATTGTCGTCAAGGTG ACCCGCTTTGATCGCGAAACAAACAAATATGAAGTGATAGACGAAGAGCCTGGGGACGAAGAAGAAAATGGTGGAACCGG TGGACCGAGGAAGTACAAGTTATCTCCCTCCGCAATCATCCAATTCCCAAAACCAGCGACAGCGCTGGATTTTCCAACGG GATCTCAAGTTCTTGCGGTGTATCCAGGCACCACCGCACTGTACAAAGCTACTGTCGTCGGTCCTCATAGAAAAAAAAAA ACGGACGACTATATTCTAGAATTCGATGACGACGACGAGGATGGATTTCTACCAAAGCGAACAGTGCCATTTTACCACGT TGTCACTCTTCCGGAGGGGCACCGCCAGTAA |