
| Microexon ID | Ps_NW_020644373.1:2755240-2755253:- |
| Species | Papaver somniferum | Coordinates | NW_020644373.1:2755240..2755253 |
| Microexon Cluster ID | MEP34 |
| Size | 14 |
| Phase | 0 |
| Pfam Domain Motif | DUF1325 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,14,46 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GGAACMACWGCACTTTAYAARGCAACTGTWGTTARTACYCMTCGMAAGAGGAARWCTGAYGAKTATTTGYTGGARTTYGATGAYGAYGARGAAGAYGGAKCTTTGCCK |
| Logo of Microexon-tag DNA Seq | 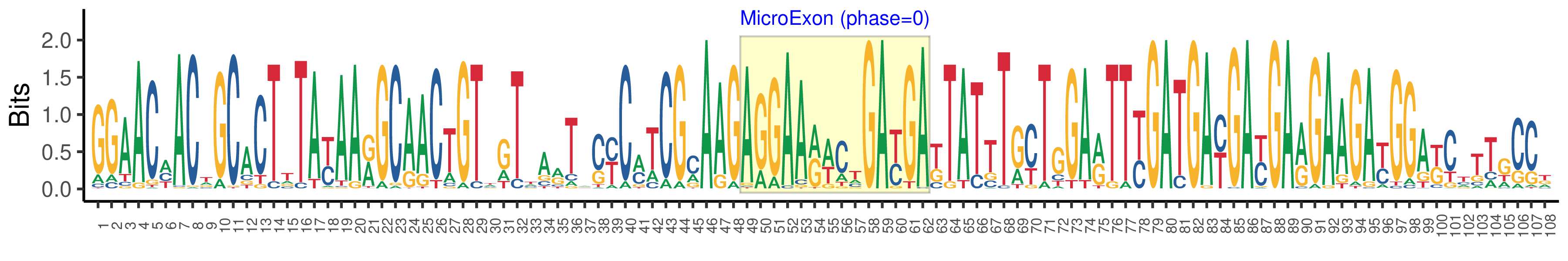 |
| Alignment of exons | 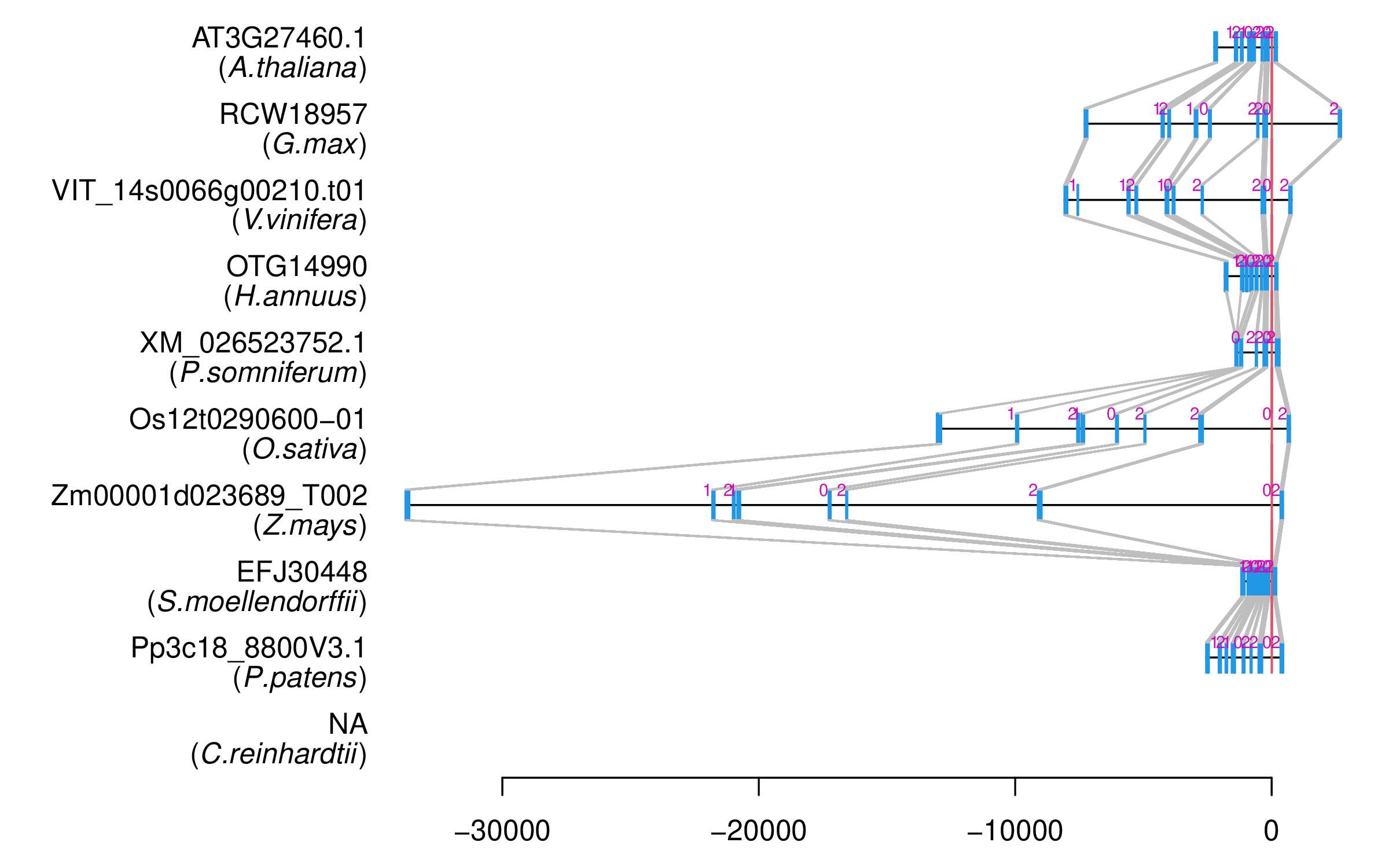 |
| Microexon DNA seq | AGGAAAAGCGACGA |
| Microexon Amino Acid seq | RKSDD |
| Microexon-tag DNA Seq | GGAACCACTGCATTATATAAAGCAACCGTGGTTAATCCTCATCGCAAGAGGAAAAGCGACGATTATTTACTCGAGTTTGATGATGATGAAGAAGATGGTTCGATGCCC |
| Microexon-tag Amino Acid Seq | GTTALYKATVVNPHRKRKSDDYLLEFDDDEEDGSMP |
| Microexon-tag spanning region | 2755047-2755466 |
| Microexon-tag prediction score | 0.9581 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | XM_026586758.1x |
| Reference Transcript ID | XM_026586758.1 |
| Gene ID | NA |
| Gene Name | NA |
| Transcript ID | XM_026586758.1 |
| Protein ID | XP_026442543.1 |
| Gene ID | LOC113342103 |
| Gene Name | NA |
| Pfam domain motif | DUF1325 |
| Motif E-value | 4e-33 |
| Motif start | 132 |
| Motif end | 266 |
| Protein seq | >XP_026442543.1 MSTIEISSILENSKELDRLRKEQEDVLLEINKIHKKLQSTPEMVEKSGDNVLLKLRALYVQAKELSENEVSVSNALINQL DTLLQSGLSAAQRKKIEVTEQKKKRMKTETDISKYPPVVSMRNQLELAAGLKGEQVAARVTPDDAEKDEWFVVKVTYFDR EAKEFEVFDEEPGDDEDGGGQRTYKLPMSRIIPFPKRNDPSSAPDYPTGKCVLAVYPGTTALYKATVVNPHRKRKSDDYL LEFDDDEEDGSMPQRTVPFHKVVPLPEGHRQ* |
| CDS seq | >XM_026586758.1 ATGTCTACGATTGAGATAAGTAGTATACTTGAGAACTCAAAGGAACTTGATCGATTAAGAAAAGAACAAGAAGATGTTCT TCTTGAAATCAATAAGATTCATAAAAAGCTTCAATCCACCCCCGAGATGGTCGAGAAGTCGGGTGATAATGTGTTATTGA AGCTCCGGGCATTGTATGTCCAAGCCAAGGAGCTTTCAGAAAATGAAGTCAGTGTCTCGAATGCACTTATAAATCAACTG GATACCTTATTACAATCAGGACTTTCTGCAGCGCAGAGGAAAAAGATAGAAGTTACTGAGCAGAAAAAGAAACGAATGAA AACTGAGACGGATATATCAAAGTATCCACCTGTTGTTTCCATGCGAAACCAACTGGAGCTTGCGGCTGGTCTCAAGGGTG AACAGGTGGCAGCTAGAGTCACACCAGATGATGCTGAGAAGGATGAGTGGTTTGTTGTAAAAGTAACTTACTTCGATAGA GAAGCAAAAGAATTTGAAGTATTTGATGAGGAACCAGGTGATGATGAAGATGGAGGCGGCCAGAGAACGTACAAGCTACC TATGTCACGCATTATTCCTTTCCCAAAGCGAAATGATCCTTCTAGTGCACCTGATTACCCAACTGGGAAATGTGTTTTGG CAGTCTATCCTGGAACCACTGCATTATATAAAGCAACCGTGGTTAATCCTCATCGCAAGAGGAAAAGCGACGATTATTTA CTCGAGTTTGATGATGATGAAGAAGATGGTTCGATGCCCCAAAGAACGGTTCCCTTCCATAAGGTGGTTCCATTGCCAGA GGGGCATCGCCAGTAG |
| Microexon DNA seq | AGGAAAAGCGACGA |
| Microexon Amino Acid seq | RKSDD |
| Microexon-tag DNA Seq | GGAACCACTGCATTATATAAAGCAACCGTGGTTAATCCTCATCGCAAGAGGAAAAGCGACGATTATTTACTCGAGTTTGATGATGATGAAGAAGATGGTTCGATGCCC |
| Microexon-tag Amino Acid seq | GTTALYKATVVNPHRKRKSDDYLLEFDDDEEDGSMP |
| Transcript ID | XM_026586758.1 |
| Gene ID | Ps.80839 |
| Gene Name | NA |
| Pfam domain motif | DUF1325 |
| Motif E-value | 4e-33 |
| Motif start | 132 |
| Motif end | 266 |
| Protein seq | >XM_026586758.1 MSTIEISSILENSKELDRLRKEQEDVLLEINKIHKKLQSTPEMVEKSGDNVLLKLRALYVQAKELSENEVSVSNALINQL DTLLQSGLSAAQRKKIEVTEQKKKRMKTETDISKYPPVVSMRNQLELAAGLKGEQVAARVTPDDAEKDEWFVVKVTYFDR EAKEFEVFDEEPGDDEDGGGQRTYKLPMSRIIPFPKRNDPSSAPDYPTGKCVLAVYPGTTALYKATVVNPHRKRKSDDYL LEFDDDEEDGSMPQRTVPFHKVVPLPEGHRQ* |
| CDS seq | >XM_026586758.1 ATGTCTACGATTGAGATAAGTAGTATACTTGAGAACTCAAAGGAACTTGATCGATTAAGAAAAGAACAAGAAGATGTTCT TCTTGAAATCAATAAGATTCATAAAAAGCTTCAATCCACCCCCGAGATGGTCGAGAAGTCGGGTGATAATGTGTTATTGA AGCTCCGGGCATTGTATGTCCAAGCCAAGGAGCTTTCAGAAAATGAAGTCAGTGTCTCGAATGCACTTATAAATCAACTG GATACCTTATTACAATCAGGACTTTCTGCAGCGCAGAGGAAAAAGATAGAAGTTACTGAGCAGAAAAAGAAACGAATGAA AACTGAGACGGATATATCAAAGTATCCACCTGTTGTTTCCATGCGAAACCAACTGGAGCTTGCGGCTGGTCTCAAGGGTG AACAGGTGGCAGCTAGAGTCACACCAGATGATGCTGAGAAGGATGAGTGGTTTGTTGTAAAAGTAACTTACTTCGATAGA GAAGCAAAAGAATTTGAAGTATTTGATGAGGAACCAGGTGATGATGAAGATGGAGGCGGCCAGAGAACGTACAAGCTACC TATGTCACGCATTATTCCTTTCCCAAAGCGAAATGATCCTTCTAGTGCACCTGATTACCCAACTGGGAAATGTGTTTTGG CAGTCTATCCTGGAACCACTGCATTATATAAAGCAACCGTGGTTAATCCTCATCGCAAGAGGAAAAGCGACGATTATTTA CTCGAGTTTGATGATGATGAAGAAGATGGTTCGATGCCCCAAAGAACGGTTCCCTTCCATAAGGTGGTTCCATTGCCAGA GGGGCATCGCCAGTAG |