
| Microexon ID | Ps_NC_039362.1:27784179-27784192:- |
| Species | Papaver somniferum | Coordinates | NC_039362.1:27784179..27784192 |
| Microexon Cluster ID | MEP34 |
| Size | 14 |
| Phase | 0 |
| Pfam Domain Motif | DUF1325 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,14,46 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GGAACMACWGCACTTTAYAARGCAACTGTWGTTARTACYCMTCGMAAGAGGAARWCTGAYGAKTATTTGYTGGARTTYGATGAYGAYGARGAAGAYGGAKCTTTGCCK |
| Logo of Microexon-tag DNA Seq | 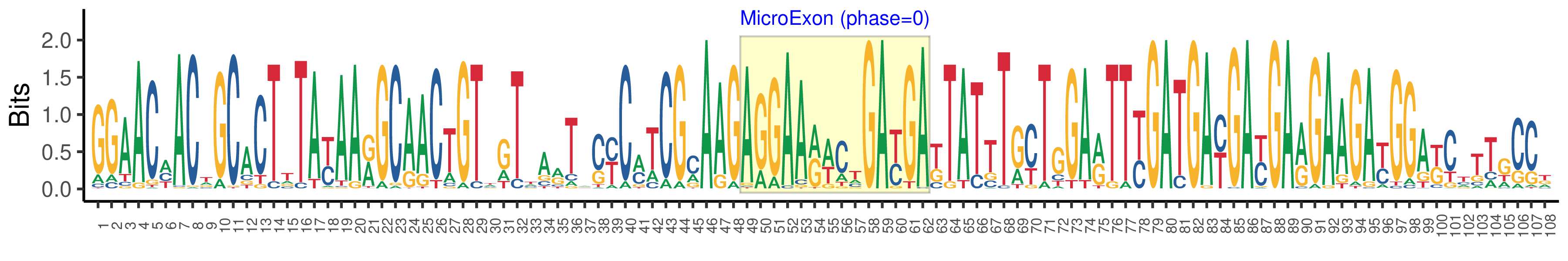 |
| Alignment of exons | 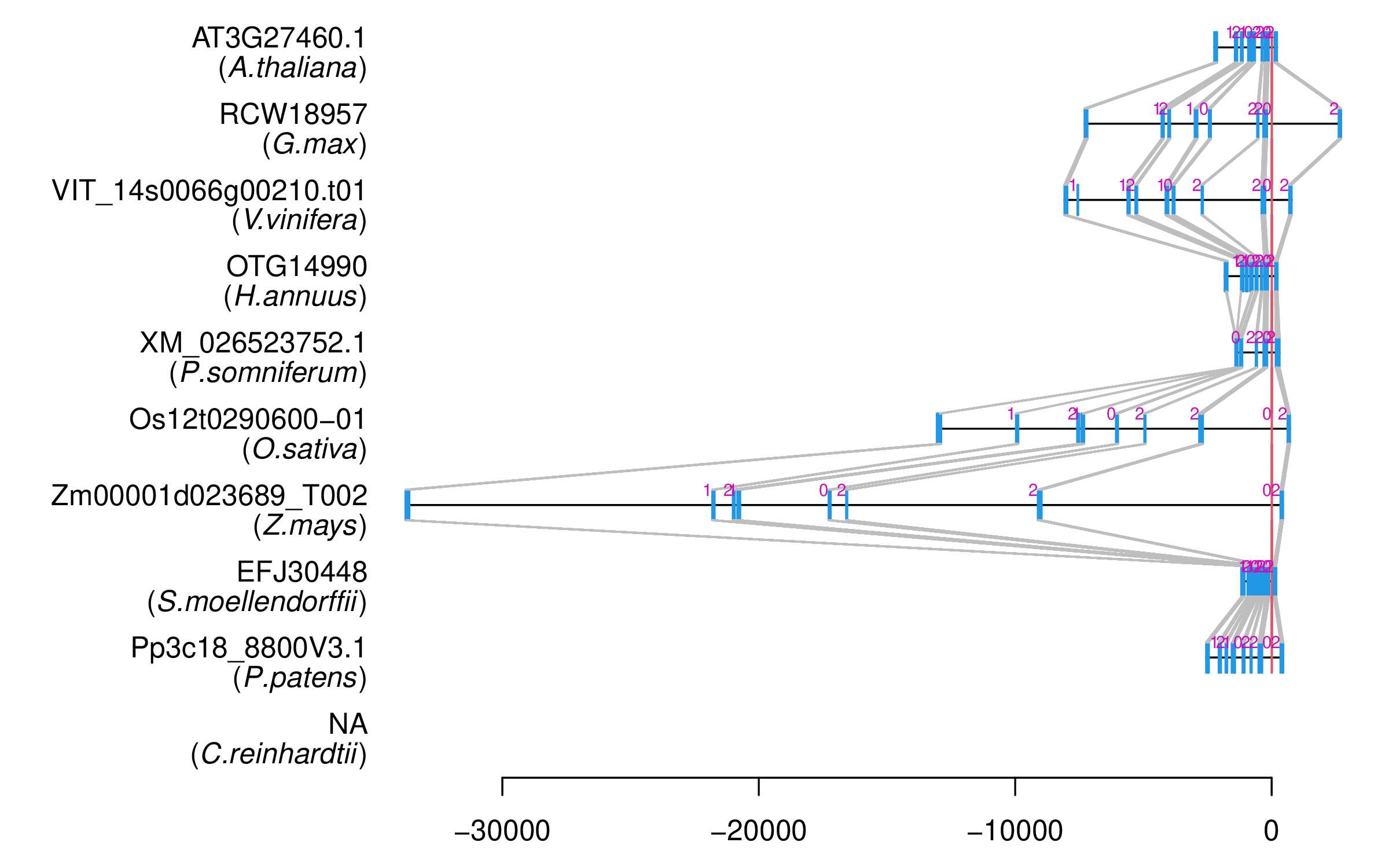 |
| Microexon DNA seq | AGGAAAAACGACGC |
| Microexon Amino Acid seq | RKNDA |
| Microexon-tag DNA Seq | GGAACCACTGCATTGTATAAAGCAACCGTGGTTAATCCTCATCGCAAGAGGAAAAACGACGCCTATTTACTGGAGTTTGATGATGATGAAGAATATCATTCGACGCCA |
| Microexon-tag Amino Acid Seq | GTTALYKATVVNPHRKRKNDAYLLEFDDDEEYHSTP |
| Microexon-tag spanning region | 27783976-27784377 |
| Microexon-tag prediction score | 0.932 |
| Overlapped with the annotated transcript (%) | NA |
| New Transcript ID | NA |
| Reference Transcript ID | NA |
| Gene ID | NA |
| Gene Name | NA |
Ps_NC_039362.1:27784179-27784192:- does not have available information here.
| Microexon DNA seq | AGGAAAAACGACGC |
| Microexon Amino Acid seq | RKNDA |
| Microexon-tag DNA Seq | GGAACCACTGCATTGTATAAAGCAACCGTGGTTAATCCTCATCGCAAGAGGAAAAACGACGCCTATTTACTGGAGTTTGATGATGATGAAGAATATCATTCGACGCCA |
| Microexon-tag Amino Acid seq | GTTALYKATVVNPHRKRKNDAYLLEFDDDEEYHSTP |
| Transcript ID | Ps.27784.1 |
| Gene ID | Ps.27784 |
| Gene Name | NA |
| Pfam domain motif | DUF1325 |
| Motif E-value | 3.5e-33 |
| Motif start | 12 |
| Motif end | 146 |
| Protein seq | >Ps.27784.1 MTNQLELAAGLKGEQVAARVTPDDAQRDEWFVVKVTHFDRETKEFEVFDVEPGDDEDGGSQRTYKLPMSRIIPFPKRNDP SNAPDYPTEKCVLAVYPGTTALYKATVVNPHRKRKNDAYLLEFDDDEEYHSTPQRTVPFHKVVPLPEVLQVMKSMSTYRL WKLLAGIRI* |
| CDS seq | >Ps.27784.1 ATGACAAACCAACTAGAGCTTGCGGCTGGTCTTAAGGGTGAACAGGTGGCAGCTAGAGTCACACCTGATGATGCTCAAAG GGATGAGTGGTTTGTTGTAAAAGTAACTCATTTCGATAGAGAAACAAAAGAATTTGAAGTATTTGATGTGGAGCCAGGTG ATGATGAAGATGGAGGCTCCCAGAGAACGTACAAGCTACCTATGTCACGGATTATTCCTTTCCCAAAGCGAAATGACCCC TCTAACGCACCTGATTACCCAACTGAGAAATGTGTTTTGGCAGTCTATCCTGGAACCACTGCATTGTATAAAGCAACCGT GGTTAATCCTCATCGCAAGAGGAAAAACGACGCCTATTTACTGGAGTTTGATGATGATGAAGAATATCATTCGACGCCAC AAAGAACGGTACCCTTCCATAAGGTGGTTCCATTGCCAGAAGTCCTACAAGTCATGAAAAGCATGTCAACATACAGATTG TGGAAATTGTTGGCCGGTATACGAATATGA |