
| Microexon ID | Ha_15:98338164-98338177:- |
| Species | Helianthus annuus | Coordinates | 15:98338164..98338177 |
| Microexon Cluster ID | MEP34 |
| Size | 14 |
| Phase | 0 |
| Pfam Domain Motif | DUF1325 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,14,46 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GGAACMACWGCACTTTAYAARGCAACTGTWGTTARTACYCMTCGMAAGAGGAARWCTGAYGAKTATTTGYTGGARTTYGATGAYGAYGARGAAGAYGGAKCTTTGCCK |
| Logo of Microexon-tag DNA Seq | 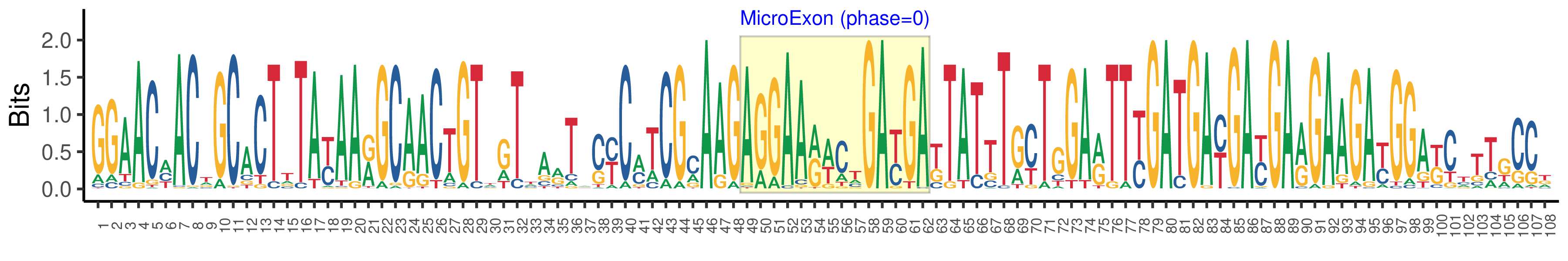 |
| Alignment of exons | 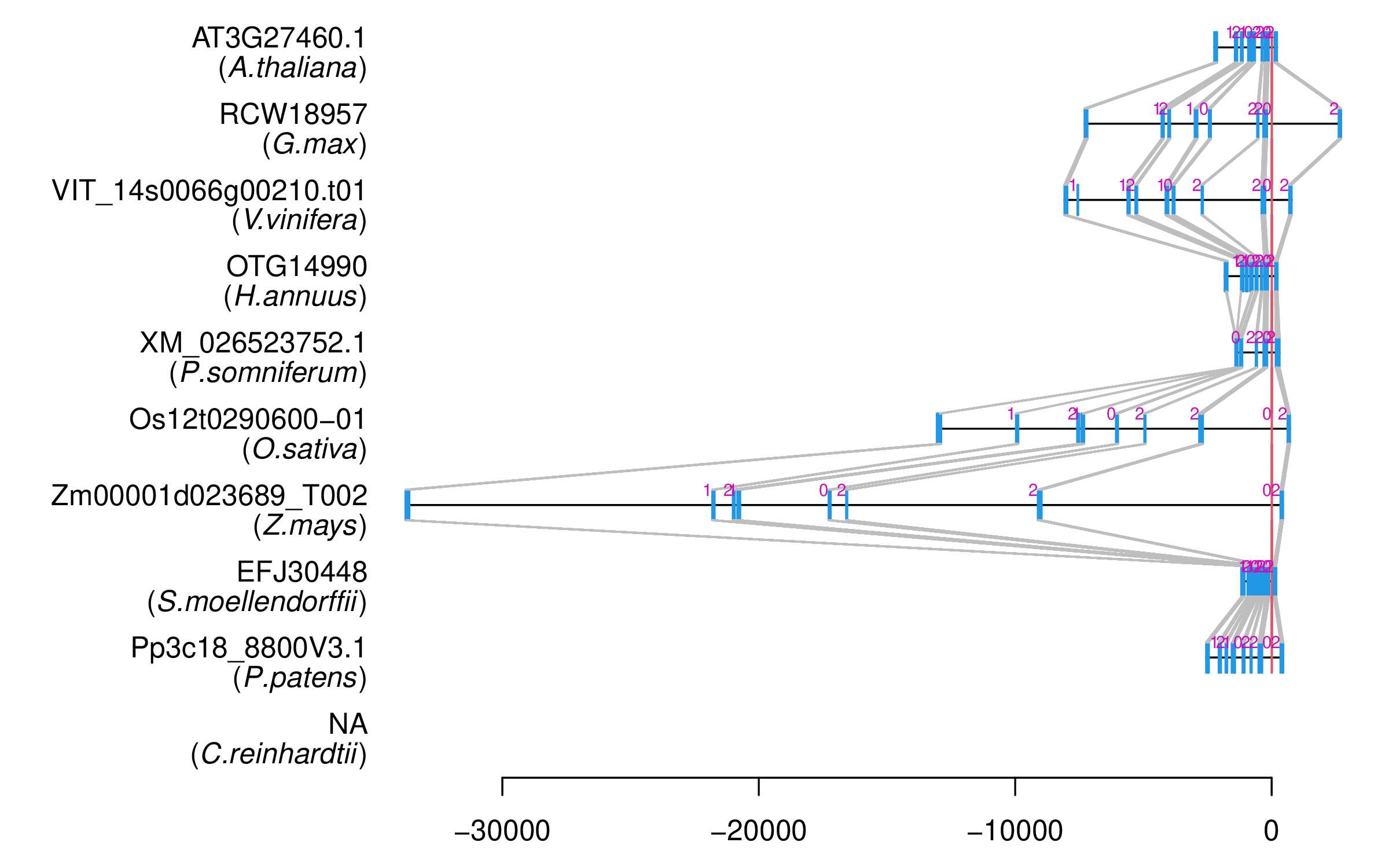 |
Ha_15:98338164-98338177:- does not have available information here.
Ha_15:98338164-98338177:- does not have available information here.
| Microexon DNA seq | AGGAAAACTGACGA |
| Microexon Amino Acid seq | RKTDD |
| Microexon-tag DNA Seq | CCTGGAACCACTGCTCTGTATAAAGCAACTGTCGTGAATACTCGCAAGAGGAAAACTGACGATTATGTTCTTGAGTTTGATGACGACGAGGAAGACGGATCCTTGCCT |
| Microexon-tag Amino Acid seq | PGTTALYKATVVNTRKRKTDDYVLEFDDDEEDGSLP |
| Transcript ID | Ha.24340.1 |
| Gene ID | Ha.24340 |
| Gene Name | NA |
| Pfam domain motif | DUF1325 |
| Motif E-value | 4.2e-36 |
| Motif start | 130 |
| Motif end | 262 |
| Protein seq | >Ha.24340.1 MSSPDVAGILDQSKELDRLRKDQEEILLEINKMHKKLQSTPEVVEKPGDNSLSRLKMLYVQAKELSENEVAVSSQLLSQL DALIPSGPLGQQRRRMEGNEQKKKRMKADPDVPRLSPSLRNHLDHLANLKGEQVAARVTQEDDDKDEWFIVKVIHFDRET REFEVLDEEPGDDEPAGQRKYKLPMSHIIPFPKRNDPSTAQDFPPGKHVLAVYPGTTALYKATVVNTRKRKTDDYVLEFD DDEEDGSLPQRHVPFHKVVTLPEGHRQ* |
| CDS seq | >Ha.24340.1 ATGTCCTCGCCGGACGTTGCTGGAATCTTGGATCAGTCAAAGGAGCTTGATCGTTTACGGAAAGATCAGGAGGAGATACT TCTTGAAATCAACAAGATGCATAAAAAGCTACAATCAACTCCTGAGGTGGTGGAGAAACCGGGTGATAATTCTCTTTCAA GGCTTAAGATGCTTTACGTTCAAGCAAAAGAACTTTCAGAAAATGAAGTAGCTGTTTCCAGTCAGTTGTTAAGTCAGTTA GATGCTCTAATACCTTCCGGGCCTCTTGGACAACAAAGAAGAAGGATGGAAGGTAATGAACAGAAGAAGAAAAGAATGAA AGCTGATCCAGATGTACCCAGGCTTTCTCCGTCTCTTCGTAATCATCTTGATCATCTTGCAAACTTAAAGGGCGAACAAG TTGCTGCTAGGGTCACACAGGAGGATGATGACAAAGACGAGTGGTTCATCGTCAAAGTAATTCACTTTGATAGGGAAACA CGAGAATTTGAAGTTCTTGATGAGGAACCAGGTGATGATGAACCAGCTGGCCAGAGAAAATACAAGCTGCCAATGTCACA TATTATACCTTTCCCGAAGCGAAATGATCCATCTACCGCTCAAGATTTCCCACCTGGAAAACACGTATTGGCAGTTTATC CTGGAACCACTGCTCTGTATAAAGCAACTGTCGTGAATACTCGCAAGAGGAAAACTGACGATTATGTTCTTGAGTTTGAT GACGACGAGGAAGACGGATCCTTGCCTCAGAGGCACGTGCCCTTCCACAAGGTAGTTACCCTTCCTGAAGGGCATCGCCA ATGA |