
| Microexon ID | Gm_19:9591565-9591578:- |
| Species | Glycine max | Coordinates | 19:9591565..9591578 |
| Microexon Cluster ID | MEP34 |
| Size | 14 |
| Phase | 0 |
| Pfam Domain Motif | DUF1325 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,14,46 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GGAACMACWGCACTTTAYAARGCAACTGTWGTTARTACYCMTCGMAAGAGGAARWCTGAYGAKTATTTGYTGGARTTYGATGAYGAYGARGAAGAYGGAKCTTTGCCK |
| Logo of Microexon-tag DNA Seq | 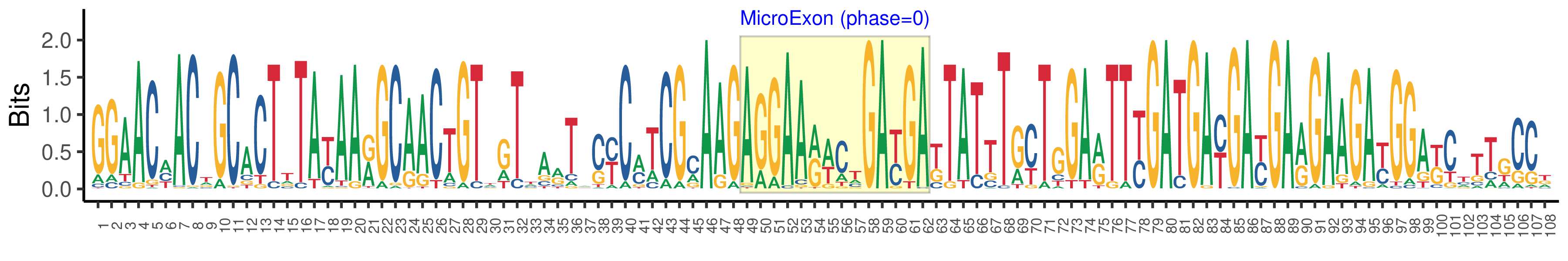 |
| Alignment of exons | 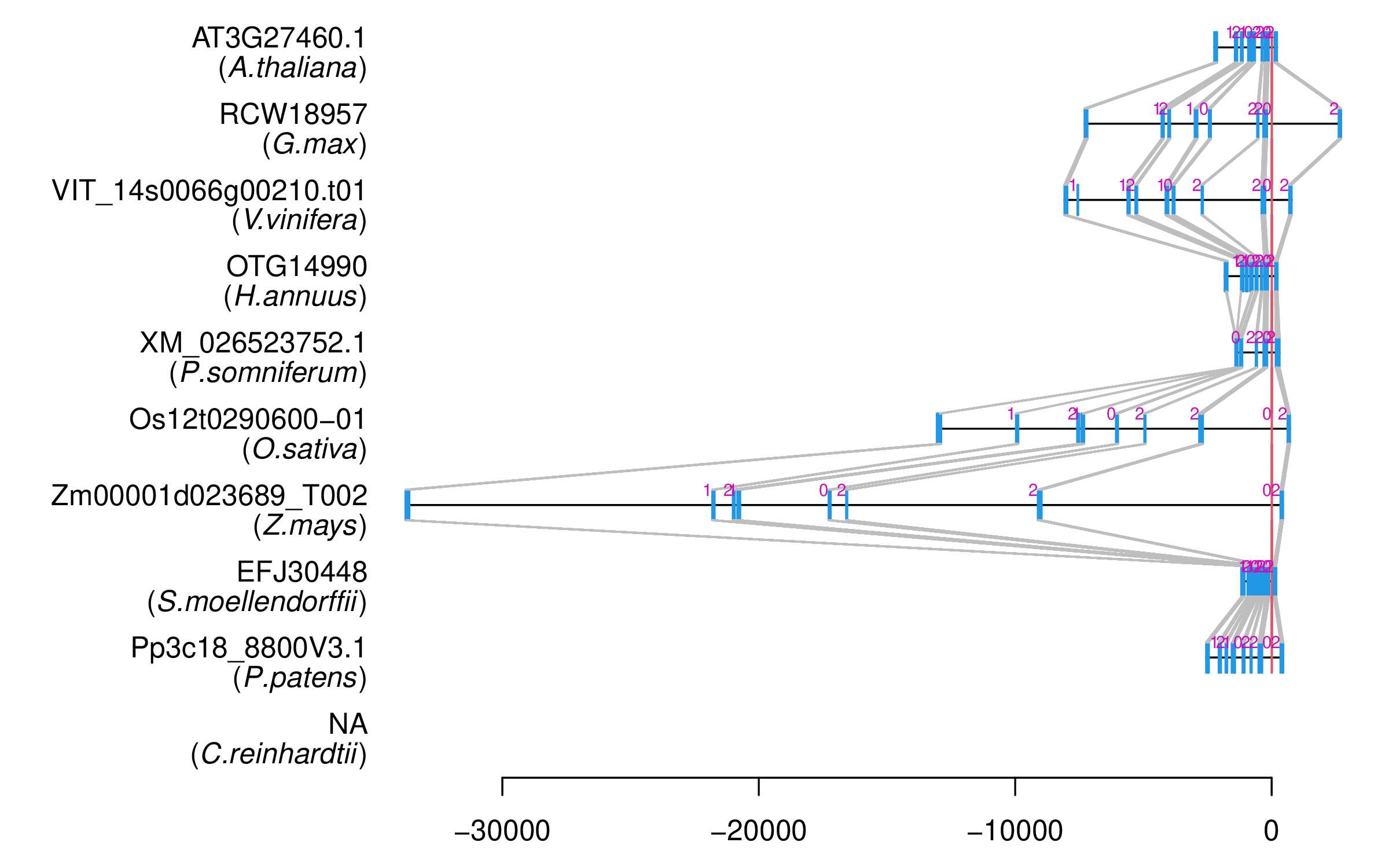 |
| Microexon DNA seq | AGGAAGACCGAGGA |
| Microexon Amino Acid seq | RKTED |
| Microexon-tag DNA Seq | GGAACTACTGCACTTTATAAAGCAACAGTTGTTCAAGGCCATCGCAGAAGGAAGACCGAGGATTACGTGTTGGAATTTGATGATGATGAGGAAGATGGATCTTTACCA |
| Microexon-tag Amino Acid Seq | GTTALYKATVVQGHRRRKTEDYVLEFDDDEEDGSLP |
| Microexon-tag spanning region | 9590257-9591804 |
| Microexon-tag prediction score | 0.9544 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRG94013x |
| Reference Transcript ID | KRG94013 |
| Gene ID | GLYMA_19G055900 |
| Gene Name | NA |
| Transcript ID | KRG94013 |
| Protein ID | KRG94013 |
| Gene ID | GLYMA_19G055900 |
| Gene Name | NA |
| Pfam domain motif | DUF1325 |
| Motif E-value | 4.8e-34 |
| Motif start | 130 |
| Motif end | 264 |
| Protein seq | >KRG94013 MSSPDIVSILDNSKELDRVRKEQEDILSEINKLHKKLQATPEVVEKPGDNSLARLKFLYTQAKELSESEASISNLLINQI DALLPTGPQGQTRRRIEGNEQKRKRVKTESDISRLTPSMRGQLEACANLKGEQVAARVTPRNADKDEWFVVKVIHFDKES KEFEVLDEEPGDDEESSGQRQYKLPMANIIPFPKSNDPSSAQDFPPGRHVLAVYPGTTALYKATVVQGHRRRKTEDYVLE FDDDEEDGSLPQRTVPFHKVVPLPEGHRQ* |
| CDS seq | >KRG94013 ATGTCGTCGCCGGACATTGTCTCGATATTGGATAACTCGAAGGAGCTCGATCGGGTTAGGAAAGAACAGGAGGATATTCT GTCGGAGATCAACAAGCTTCACAAGAAGCTTCAAGCAACTCCTGAAGTGGTTGAGAAGCCTGGGGATAACTCATTGGCAA GGCTGAAATTTTTGTATACTCAAGCAAAAGAACTTTCAGAAAGTGAAGCAAGTATTTCCAACTTACTGATAAACCAAATT GATGCCCTACTGCCTACGGGTCCACAAGGGCAAACACGAAGAAGAATAGAAGGTAATGAGCAGAAAAGGAAACGAGTGAA GACTGAGTCAGATATTTCAAGGCTAACTCCTAGTATGCGAGGTCAACTTGAGGCTTGTGCCAATCTTAAAGGTGAACAGG TAGCAGCTAGGGTCACACCACGAAATGCCGATAAGGATGAATGGTTTGTCGTAAAAGTGATTCATTTTGATAAGGAATCA AAGGAATTTGAAGTACTAGATGAGGAACCAGGTGATGATGAAGAAAGCAGTGGCCAAAGACAATACAAGCTTCCCATGGC AAATATCATTCCTTTTCCCAAGAGTAATGATCCTTCTAGCGCTCAAGATTTCCCTCCTGGAAGACATGTTTTGGCGGTCT ATCCAGGAACTACTGCACTTTATAAAGCAACAGTTGTTCAAGGCCATCGCAGAAGGAAGACCGAGGATTACGTGTTGGAA TTTGATGATGATGAGGAAGATGGATCTTTACCACAAAGGACAGTGCCCTTCCACAAAGTGGTTCCTCTTCCAGAAGGACA TCGCCAATGA |
| Microexon DNA seq | AGGAAGACCGAGGA |
| Microexon Amino Acid seq | RKTED |
| Microexon-tag DNA Seq | GGAACTACTGCACTTTATAAAGCAACAGTTGTTCAAGGCCATCGCAGAAGGAAGACCGAGGATTACGTGTTGGAATTTGATGATGATGAGGAAGATGGATCTTTACCA |
| Microexon-tag Amino Acid seq | GTTALYKATVVQGHRRRKTEDYVLEFDDDEEDGSLP |
| Transcript ID | KRG94013 |
| Gene ID | Gm.27294 |
| Gene Name | NA |
| Pfam domain motif | DUF1325 |
| Motif E-value | 4.9e-34 |
| Motif start | 130 |
| Motif end | 264 |
| Protein seq | >KRG94013 MSSPDIVSILDNSKELDRVRKEQEDILSEINKLHKKLQATPEVVEKPGDNSLARLKFLYTQAKELSESEASISNLLINQI DALLPTGPQGQTRRRIEGNEQKRKRVKTESDISRLTPSMRGQLEACANLKGEQVAARVTPRNADKDEWFVVKVIHFDKES KEFEVLDEEPGDDEESSGQRQYKLPMANIIPFPKSNDPSSAQDFPPGRHVLAVYPGTTALYKATVVQGHRRRKTEDYVLE FDDDEEDGSLPQRTVPFHKVVPLPEGHRQ* |
| CDS seq | >KRG94013 ATGTCGTCGCCGGACATTGTCTCGATATTGGATAACTCGAAGGAGCTCGATCGGGTTAGGAAAGAACAGGAGGATATTCT GTCGGAGATCAACAAGCTTCACAAGAAGCTTCAAGCAACTCCTGAAGTGGTTGAGAAGCCTGGGGATAACTCATTGGCAA GGCTGAAATTTTTGTATACTCAAGCAAAAGAACTTTCAGAAAGTGAAGCAAGTATTTCCAACTTACTGATAAACCAAATT GATGCCCTACTGCCTACGGGTCCACAAGGGCAAACACGAAGAAGAATAGAAGGTAATGAGCAGAAAAGGAAACGAGTGAA GACTGAGTCAGATATTTCAAGGCTAACTCCTAGTATGCGAGGTCAACTTGAGGCTTGTGCCAATCTTAAAGGTGAACAGG TAGCAGCTAGGGTCACACCACGAAATGCCGATAAGGATGAATGGTTTGTCGTAAAAGTGATTCATTTTGATAAGGAATCA AAGGAATTTGAAGTACTAGATGAGGAACCAGGTGATGATGAAGAAAGCAGTGGCCAAAGACAATACAAGCTTCCCATGGC AAATATCATTCCTTTTCCCAAGAGTAATGATCCTTCTAGCGCTCAAGATTTCCCTCCTGGAAGACATGTTTTGGCGGTCT ATCCAGGAACTACTGCACTTTATAAAGCAACAGTTGTTCAAGGCCATCGCAGAAGGAAGACCGAGGATTACGTGTTGGAA TTTGATGATGATGAGGAAGATGGATCTTTACCACAAAGGACAGTGCCCTTCCACAAAGTGGTTCCTCTTCCAGAAGGACA TCGCCAATGA |