
| Microexon ID | At_5:16242896-16242909:+ |
| Species | Arabidopsis thaliana | Coordinates | 5:16242896..16242909 |
| Microexon Cluster ID | MEP34 |
| Size | 14 |
| Phase | 0 |
| Pfam Domain Motif | DUF1325 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,14,46 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GGAACMACWGCACTTTAYAARGCAACTGTWGTTARTACYCMTCGMAAGAGGAARWCTGAYGAKTATTTGYTGGARTTYGATGAYGAYGARGAAGAYGGAKCTTTGCCK |
| Logo of Microexon-tag DNA Seq | 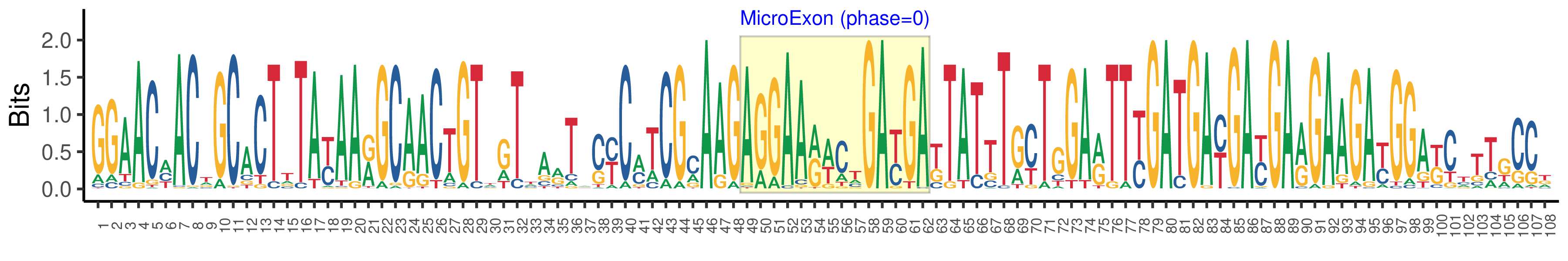 |
| Alignment of exons | 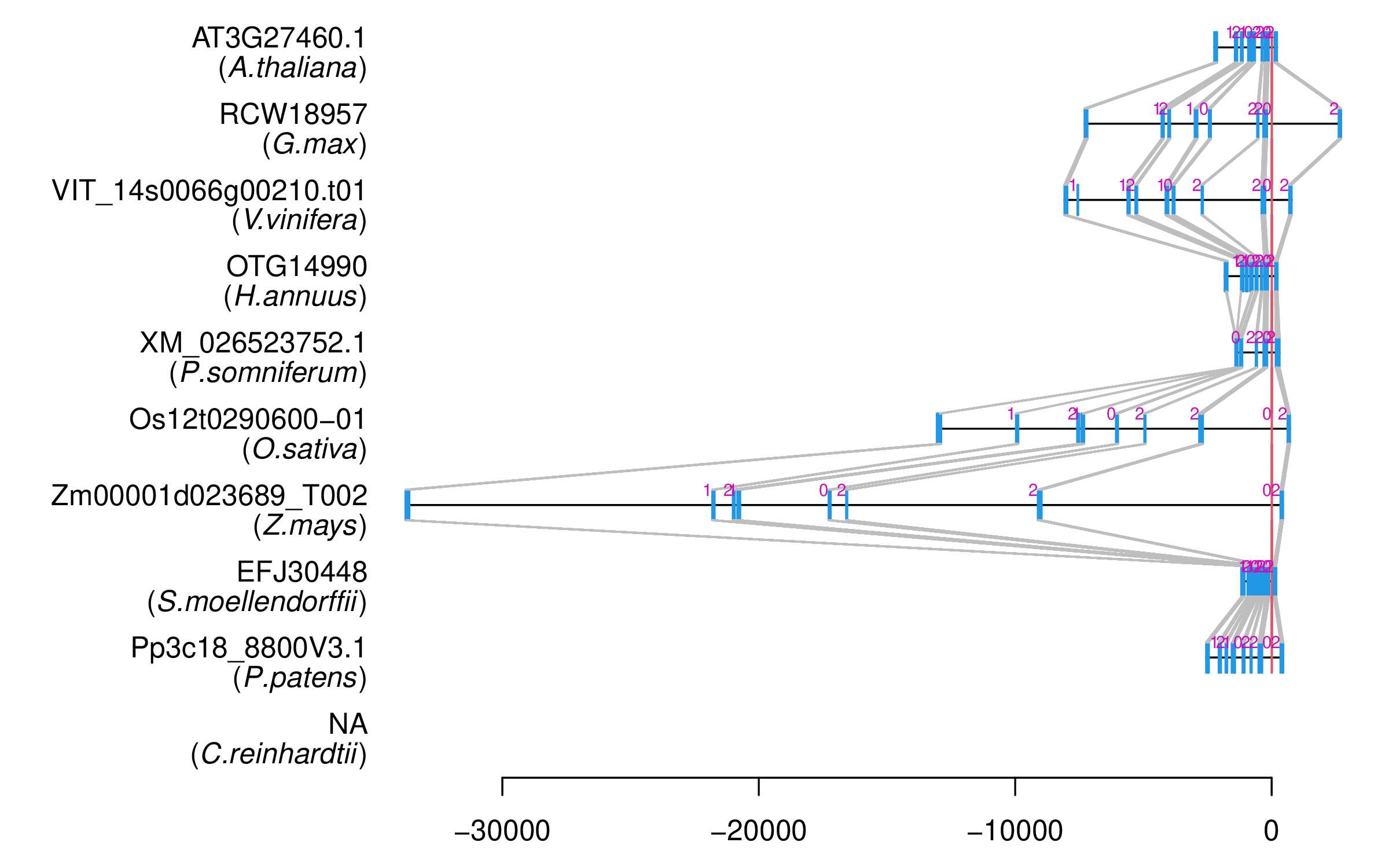 |
| Microexon DNA seq | AGGAAGTCTGACGA |
| Microexon Amino Acid seq | RKSDE |
| Microexon-tag DNA Seq | GGGACAACAGCACTCTACAAAGCAACTGTAGTAAGTACCCCACGAAAGAGGAAGTCTGACGAGTATTTGCTTGAATTCGACGATGATGAGGAAGATGGAGCATTGCCT |
| Microexon-tag Amino Acid Seq | GTTALYKATVVSTPRKRKSDEYLLEFDDDEEDGALP |
| Microexon-tag spanning region | 16242738-16243043 |
| Microexon-tag prediction score | 0.9711 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | AT5G40550.1x |
| Reference Transcript ID | AT5G40550.1 |
| Gene ID | AT5G40550 |
| Gene Name | NA |
| Transcript ID | AT5G40550.1 |
| Protein ID | AT5G40550.1 |
| Gene ID | AT5G40550 |
| Gene Name | NA |
| Pfam domain motif | DUF1325 |
| Motif E-value | 2.7e-32 |
| Motif start | 134 |
| Motif end | 268 |
| Protein seq | >AT5G40550.1 MSSPDIVGILENTKELDRLRKDQEEVLVEINKMHKKLQASPEIVEKPGDISLAKLKNLYIQAKELSENEVTVSNILLTQL DLLLPYGPTGQQRRKLGVVAEGNDQKRKRMKVDSDVIRLSPSMRNQIEAYASLKGEQVAARVTAESADKDEWFVVKVIHF DRETKEVEVLDEEPGDDEEGSGQRTYKLPMLCILPFPKRNDPSNTQEFPPGKHVLAVYPGTTALYKATVVSTPRKRKSDE YLLEFDDDEEDGALPQRTVPFHKVVALPEGHRQ* |
| CDS seq | >AT5G40550.1 ATGTCGTCACCAGACATTGTCGGAATTTTGGAAAACACGAAGGAACTTGATCGGTTAAGGAAAGACCAAGAAGAAGTTTT GGTTGAGATCAATAAGATGCATAAAAAGCTTCAAGCTTCTCCTGAGATAGTTGAGAAGCCTGGTGATATTTCATTAGCAA AGCTTAAGAATTTGTACATTCAAGCGAAAGAGCTCTCAGAAAATGAAGTAACCGTATCGAACATTTTGCTTACTCAGTTG GATTTGCTGCTTCCATATGGACCAACAGGGCAGCAAAGAAGAAAGTTAGGCGTGGTGGCAGAAGGTAATGATCAGAAGAG GAAGAGGATGAAAGTTGATTCAGATGTAATAAGACTTTCTCCTTCCATGAGAAATCAAATCGAAGCATATGCAAGTCTTA AGGGTGAACAGGTTGCTGCAAGAGTTACTGCAGAGAGTGCTGATAAGGATGAATGGTTTGTGGTTAAAGTTATTCACTTC GATAGAGAAACAAAAGAAGTTGAAGTACTTGATGAAGAACCAGGAGATGATGAAGAAGGAAGCGGTCAGAGGACCTATAA GTTGCCAATGTTATGCATTTTACCATTCCCAAAGCGAAACGATCCTTCGAACACCCAAGAGTTTCCACCGGGTAAACACG TCTTAGCTGTATATCCCGGGACAACAGCACTCTACAAAGCAACTGTAGTAAGTACCCCACGAAAGAGGAAGTCTGACGAG TATTTGCTTGAATTCGACGATGATGAGGAAGATGGAGCATTGCCTCAAAGAACAGTACCTTTCCACAAAGTGGTAGCTTT ACCAGAAGGTCATCGCCAGTGA |
| Microexon DNA seq | AGGAAGTCTGACGA |
| Microexon Amino Acid seq | RKSDE |
| Microexon-tag DNA Seq | GGGACAACAGCACTCTACAAAGCAACTGTAGTAAGTACCCCACGAAAGAGGAAGTCTGACGAGTATTTGCTTGAATTCGACGATGATGAGGAAGATGGAGCATTGCCT |
| Microexon-tag Amino Acid seq | GTTALYKATVVSTPRKRKSDEYLLEFDDDEEDGALP |
| Transcript ID | AT5G40550.2 |
| Gene ID | At.25045 |
| Gene Name | NA |
| Pfam domain motif | DUF1325 |
| Motif E-value | 2.6e-32 |
| Motif start | 131 |
| Motif end | 265 |
| Protein seq | >AT5G40550.2 MSSPDIVGILENTKELDRLRKDQEEVLVEINKMHKKLQASPEIVEKPGDISLAKLKNLYIQAKELSENEVTVSNILLTQL DLLLPYGPTGQQRRKLGEGNDQKRKRMKVDSDVIRLSPSMRNQIEAYASLKGEQVAARVTAESADKDEWFVVKVIHFDRE TKEVEVLDEEPGDDEEGSGQRTYKLPMLCILPFPKRNDPSNTQEFPPGKHVLAVYPGTTALYKATVVSTPRKRKSDEYLL EFDDDEEDGALPQRTVPFHKVVALPEGHRQ* |
| CDS seq | >AT5G40550.2 ATGTCGTCACCAGACATTGTCGGAATTTTGGAAAACACGAAGGAACTTGATCGGTTAAGGAAAGACCAAGAAGAAGTTTT GGTTGAGATCAATAAGATGCATAAAAAGCTTCAAGCTTCTCCTGAGATAGTTGAGAAGCCTGGTGATATTTCATTAGCAA AGCTTAAGAATTTGTACATTCAAGCGAAAGAGCTCTCAGAAAATGAAGTAACCGTATCGAACATTTTGCTTACTCAGTTG GATTTGCTGCTTCCATATGGACCAACAGGGCAGCAAAGAAGAAAGTTAGGCGAAGGTAATGATCAGAAGAGGAAGAGGAT GAAAGTTGATTCAGATGTAATAAGACTTTCTCCTTCCATGAGAAATCAAATCGAAGCATATGCAAGTCTTAAGGGTGAAC AGGTTGCTGCAAGAGTTACTGCAGAGAGTGCTGATAAGGATGAATGGTTTGTGGTTAAAGTTATTCACTTCGATAGAGAA ACAAAAGAAGTTGAAGTACTTGATGAAGAACCAGGAGATGATGAAGAAGGAAGCGGTCAGAGGACCTATAAGTTGCCAAT GTTATGCATTTTACCATTCCCAAAGCGAAACGATCCTTCGAACACCCAAGAGTTTCCACCGGGTAAACACGTCTTAGCTG TATATCCCGGGACAACAGCACTCTACAAAGCAACTGTAGTAAGTACCCCACGAAAGAGGAAGTCTGACGAGTATTTGCTT GAATTCGACGATGATGAGGAAGATGGAGCATTGCCTCAAAGAACAGTACCTTTCCACAAAGTGGTAGCTTTACCAGAAGG TCATCGCCAGTGA |