
| Microexon ID | Zm_2:197850277-197850289:+ |
| Species | Zea mays | Coordinates | 2:197850277..197850289 |
| Microexon Cluster ID | MEP32 |
| Size | 13 |
| Phase | 0 |
| Pfam Domain Motif | MCM6_C |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,13,47 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | AMYRTTGAWSTKWCWGATGTYCTYTCTARYTTCCCKGACATMTCARTGGHWCTGRYTGAAGAWATYATGGAKARRCTWSTWAAMSAWRRTRTACTRTCAARRRCRGGA |
| Logo of Microexon-tag DNA Seq | 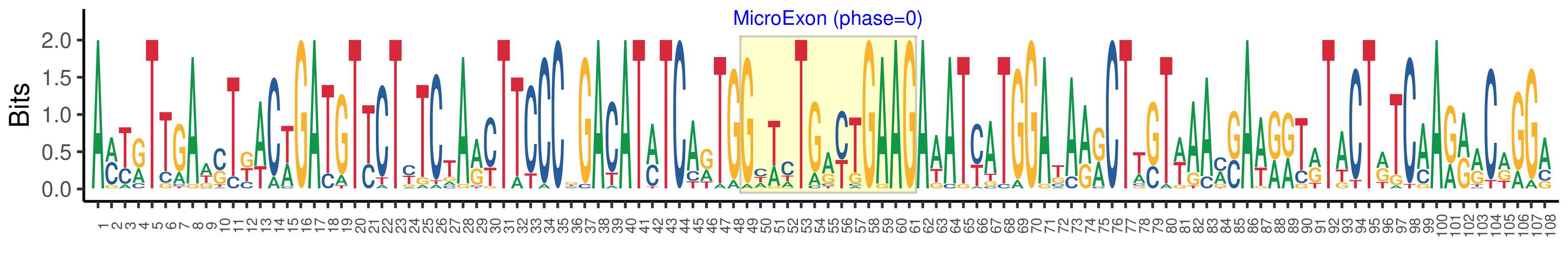 |
| Alignment of exons | 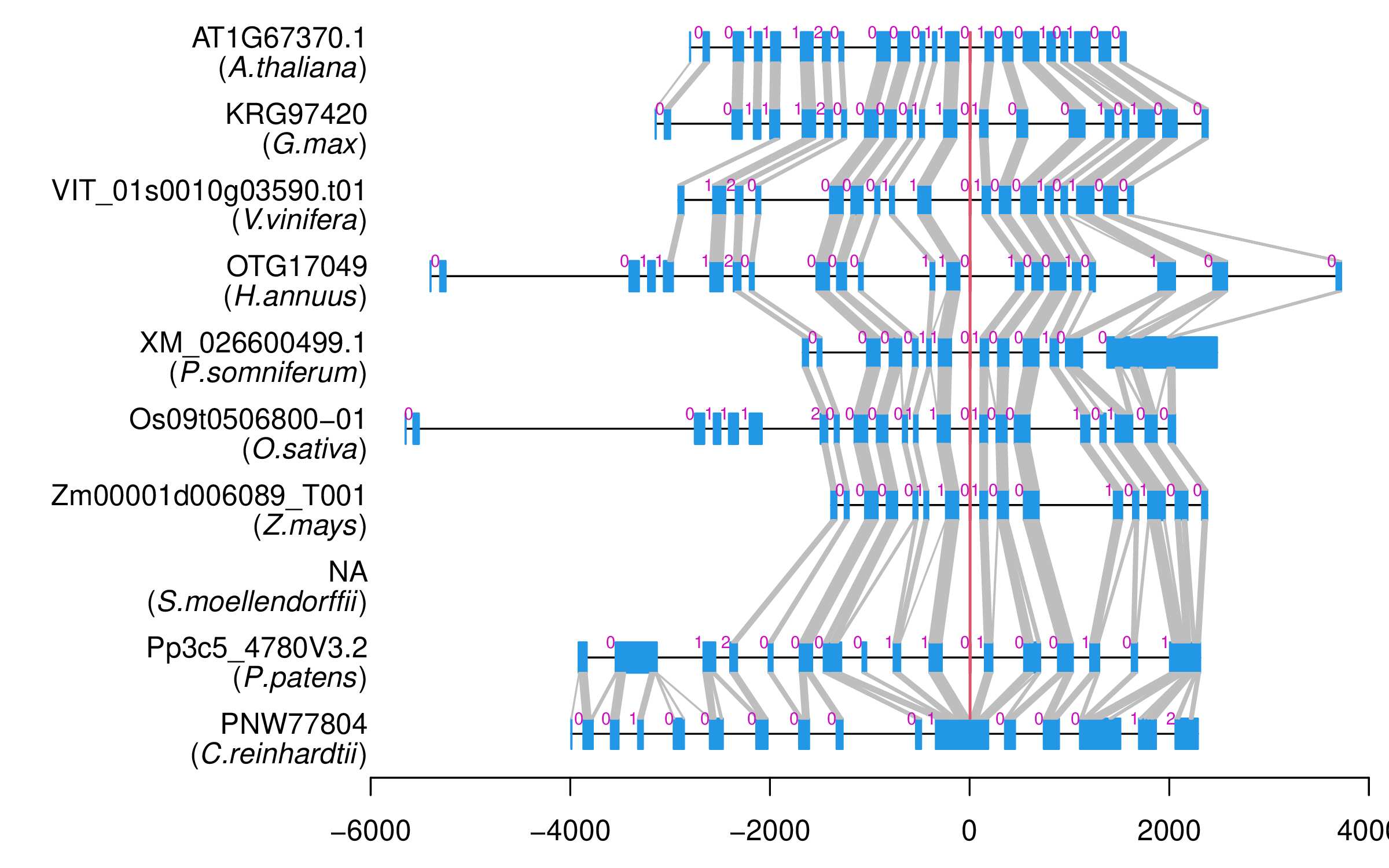 |
| Microexon DNA seq | GAATTGGTTGAAG |
| Microexon Amino Acid seq | ELVED |
| Microexon-tag DNA Seq | ACAATTAATGTTTCAGATGTCCTTTCTAACTTCCCTGACATATCAATGGAATTGGTTGAAGATATAATGGACAGGCTACTTAAAGATGGTATACTATCCAGGGCAAGC |
| Microexon-tag Amino Acid Seq | TINVSDVLSNFPDISMELVEDIMDRLLKDGILSRAS |
| Microexon-tag spanning region | 197850118-197850430 |
| Microexon-tag prediction score | 0.9434 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | Zm00001d006089_T001x |
| Reference Transcript ID | Zm00001d006089_T001 |
| Gene ID | Zm00001d006089 |
| Gene Name | NA |
| Transcript ID | Zm00001d006089_T001 |
| Protein ID | Zm00001d006089_P001 |
| Gene ID | Zm00001d006089 |
| Gene Name | NA |
| Pfam domain motif | Unknown |
| Motif E-value | NA |
| Motif start | NA |
| Motif end | NA |
| Protein seq | >Zm00001d006089_P001 MIRTLVSLMRTLDPMPEERTILMKLLYYDDVTPEDYEPPFFKCCADNEAISIWNKNPLKMEVGNVNSKHLVLALKVKSVL DPCDDDNVNSEGDGMTVDNESDHDDDFSDTEVHPPKADHYIIAPNEGKDKDQDTGTISEDDTQDTAHEEELTSQVKEWIC SRDSGTINVSDVLSNFPDISMELVEDIMDRLLKDGILSRASKDGYTVNQKADPKTPHIKKEAIMPDVSPTNVTKHNNGDL LYMKALYHALPMDYVTIAKLQGKLDGEASQNTVRKLIDKMVQDGYVKNSANRRLGKAVIHSESTNKRLLEIKKILEVNEG EKMAIDTNTEHPEPEHKDLLRVYEVGDGSTMGCLHSIGSDLTRTRELPELQQNVSMQSGQEASTVGKDSSRTPTSVRELA VPVCSLESGVLGKRIKRSLTGGNEMQSTQDKRSRKASMVKEAILQHVKCQNPQAQ* |
| CDS seq | >Zm00001d006089_T001 ATGATCAGAACGCTGGTTTCACTTATGAGAACCTTGGACCCAATGCCTGAGGAGCGAACCATTCTAATGAAGCTTCTATA CTATGATGATGTCACACCCGAGGATTATGAGCCTCCTTTCTTCAAGTGTTGTGCTGATAATGAGGCCATAAGTATATGGA ACAAGAACCCCTTGAAGATGGAAGTGGGCAATGTCAATAGCAAGCACCTTGTGTTAGCTTTGAAGGTCAAGAGTGTCCTT GATCCATGCGATGATGACAATGTTAACAGCGAAGGTGATGGCATGACTGTGGATAATGAGTCTGATCATGATGATGACTT TTCTGACACTGAGGTTCATCCACCAAAGGCGGATCATTACATCATAGCTCCTAATGAAGGAAAAGACAAAGATCAGGACA CTGGTACTATTTCAGAAGATGATACTCAAGATACTGCTCATGAGGAAGAGCTAACTTCCCAAGTGAAGGAGTGGATATGC TCAAGGGACAGCGGAACAATTAATGTTTCAGATGTCCTTTCTAACTTCCCTGACATATCAATGGAATTGGTTGAAGATAT AATGGACAGGCTACTTAAAGATGGTATACTATCCAGGGCAAGCAAAGATGGTTATACGGTTAACCAGAAGGCTGATCCAA AAACACCACATATAAAGAAGGAGGCCATCATGCCAGATGTATCACCAACTAATGTAACCAAACACAACAATGGGGATCTG CTATATATGAAGGCATTATATCATGCACTTCCCATGGATTATGTGACAATAGCCAAACTTCAAGGAAAGCTGGATGGGGA AGCCAGCCAAAATACAGTCCGAAAGTTGATTGACAAGATGGTGCAAGATGGATATGTGAAGAATTCAGCCAACCGAAGAC TGGGCAAAGCTGTCATTCATTCTGAATCTACCAACAAAAGGCTCCTTGAGATAAAAAAGATACTGGAAGTCAATGAAGGC GAAAAGATGGCCATTGACACCAATACAGAGCATCCTGAGCCTGAGCACAAAGACCTTCTGAGAGTCTATGAAGTGGGAGA TGGCTCCACTATGGGTTGCCTCCACTCAATTGGATCTGATCTTACTCGTACTCGGGAGCTGCCAGAGCTGCAACAGAATG TGTCCATGCAGAGCGGGCAAGAAGCTTCCACAGTGGGTAAGGATTCCAGCAGGACTCCAACAAGTGTGCGTGAGCTGGCT GTACCTGTTTGTTCCCTGGAAAGTGGAGTGCTTGGAAAGAGAATCAAAAGGTCTCTAACTGGTGGAAATGAGATGCAGTC TACCCAGGACAAGCGATCCAGGAAGGCTAGCATGGTGAAGGAAGCAATACTCCAACATGTCAAGTGCCAAAACCCTCAGG CTCAGTGA |
| Microexon DNA seq | GAATTGGTTGAAG |
| Microexon Amino Acid seq | ELVED |
| Microexon-tag DNA Seq | ACAATTAATGTTTCAGATGTCCTTTCTAACTTCCCTGACATATCAATGGAATTGGTTGAAGATATAATGGACAGGCTACTTAAAGATGGTATACTATCCAGGGCAAGC |
| Microexon-tag Amino Acid seq | TINVSDVLSNFPDISMELVEDIMDRLLKDGILSRAS |
| Transcript ID | Zm.11801.1 |
| Gene ID | Zm.11801 |
| Gene Name | NA |
| Pfam domain motif | Unknown |
| Motif E-value | NA |
| Motif start | NA |
| Motif end | NA |
| Protein seq | >Zm.11801.1 MAQKTKEAEITEQDSLLLTRNLLRIAIYNISYIRGLFHEKYFSDKSVPALEMKIKKLMPMDAESRRLIDWMEKGVYDALQ KKYLKTLLFCICEKEEGPMIEEYAFSFSYPNTSTEEVAMNMSRTGSKKGSTTFTSNASEVTPDQMRSSACKMIRTLVSLM RTLDPMPEERTILMKLLYYDDVTPEDYEPPFFKCCADNEAISIWNKNPLKMEVGNVNSKHLVLALKVKSVLDPCDDDNVN SEGDGMTVDNESDHDDDFSDTEVHPPKADHYIIAPNEGKDKDQDTGTISEDDTQDTAHEEELTSQVKEWICSRDSGTINV SDVLSNFPDISMELVEDIMDRLLKDGILSRASKDGYTVNQKADPKTPHIKKEAIMPDVSPTNVTKHNNGDLLYMKALYHA LPMDYVTIAKLQGKLDGEASQNTVRKLIDKMVQDGYVKNSANRRLGKAVIHSESTNKRLLEIKKILEVNEGEKMAIDTNT EHPEPEHKDLLRVYEVGDGSTMGCLHSIGSDLTRTRELPELQQNVSMQSGQEASTVGKDSSRTPTSVRELAVPVCSLESG VLGKRIKRSLTGGNEMQSTQDKRSRKASMVKEAILQHVKCQNPQAQ* |
| CDS seq | >Zm.11801.1 ATGGCTCAGAAGACGAAGGAGGCGGAGATCACCGAGCAGGATTCGCTGCTTCTAACAAGGAATTTGCTCCGGATTGCCAT ATACAACATCAGCTACATCAGAGGCCTATTCCATGAGAAGTACTTCAGTGATAAGTCGGTTCCAGCACTAGAGATGAAGA TTAAGAAGCTGATGCCAATGGATGCTGAGTCAAGGAGACTAATTGATTGGATGGAGAAAGGTGTCTATGATGCTTTGCAA AAGAAATATCTCAAGACCCTCCTCTTCTGTATCTGCGAGAAGGAGGAGGGCCCAATGATTGAGGAGTATGCCTTCTCATT TAGCTACCCCAACACCAGTACCGAGGAAGTTGCGATGAACATGAGCCGTACGGGGAGCAAAAAGGGCAGTACTACATTCA CTTCAAATGCTTCAGAAGTAACTCCTGATCAGATGAGAAGCTCTGCTTGTAAGATGATCAGAACGCTGGTTTCACTTATG AGAACCTTGGACCCAATGCCTGAGGAGCGAACCATTCTAATGAAGCTTCTATACTATGATGATGTCACACCCGAGGATTA TGAGCCTCCTTTCTTCAAGTGTTGTGCTGATAATGAGGCCATAAGTATATGGAACAAGAACCCCTTGAAGATGGAAGTGG GCAATGTCAATAGCAAGCACCTTGTGTTAGCTTTGAAGGTCAAGAGTGTCCTTGATCCATGCGATGATGACAATGTTAAC AGCGAAGGTGATGGCATGACTGTGGATAATGAGTCTGATCATGATGATGACTTTTCTGACACTGAGGTTCATCCACCAAA GGCGGATCATTACATCATAGCTCCTAATGAAGGAAAAGACAAAGATCAGGACACTGGTACTATTTCAGAAGATGATACTC AAGATACTGCTCATGAGGAAGAGCTAACTTCCCAAGTGAAGGAGTGGATATGCTCAAGGGACAGCGGAACAATTAATGTT TCAGATGTCCTTTCTAACTTCCCTGACATATCAATGGAATTGGTTGAAGATATAATGGACAGGCTACTTAAAGATGGTAT ACTATCCAGGGCAAGCAAAGATGGTTATACGGTTAACCAGAAGGCTGATCCAAAAACACCACATATAAAGAAGGAGGCCA TCATGCCAGATGTATCACCAACTAATGTAACCAAACACAACAATGGGGATCTGCTATATATGAAGGCATTATATCATGCA CTTCCCATGGATTATGTGACAATAGCCAAACTTCAAGGAAAGCTGGATGGGGAAGCCAGCCAAAATACAGTCCGAAAGTT GATTGACAAGATGGTGCAAGATGGATATGTGAAGAATTCAGCCAACCGAAGACTGGGCAAAGCTGTCATTCATTCTGAAT CTACCAACAAAAGGCTCCTTGAGATAAAAAAGATACTGGAAGTCAATGAAGGCGAAAAGATGGCCATTGACACCAATACA GAGCATCCTGAGCCTGAGCACAAAGACCTTCTGAGAGTCTATGAAGTGGGAGATGGCTCCACTATGGGTTGCCTCCACTC AATTGGATCTGATCTTACTCGTACTCGGGAGCTGCCAGAGCTGCAACAGAATGTGTCCATGCAGAGCGGGCAAGAAGCTT CCACAGTGGGTAAGGATTCCAGCAGGACTCCAACAAGTGTGCGTGAGCTGGCTGTACCTGTTTGTTCCCTGGAAAGTGGA GTGCTTGGAAAGAGAATCAAAAGGTCTCTAACTGGTGGAAATGAGATGCAGTCTACCCAGGACAAGCGATCCAGGAAGGC TAGCATGGTGAAGGAAGCAATACTCCAACATGTCAAGTGCCAAAACCCTCAGGCTCAGTGA |