
| Microexon ID | Ps_NC_039365.1:18352680-18352692:- |
| Species | Papaver somniferum | Coordinates | NC_039365.1:18352680..18352692 |
| Microexon Cluster ID | MEP32 |
| Size | 13 |
| Phase | 0 |
| Pfam Domain Motif | MCM6_C |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,13,47 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | AMYRTTGAWSTKWCWGATGTYCTYTCTARYTTCCCKGACATMTCARTGGHWCTGRYTGAAGAWATYATGGAKARRCTWSTWAAMSAWRRTRTACTRTCAARRRCRGGA |
| Logo of Microexon-tag DNA Seq | 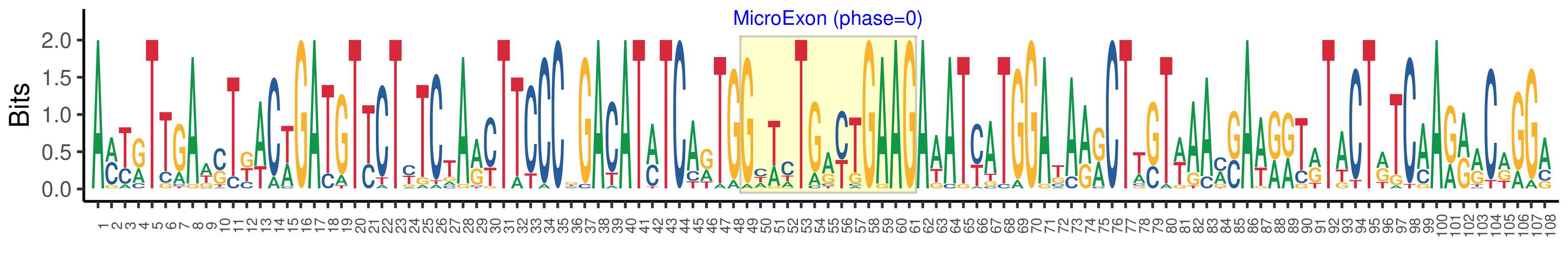 |
| Alignment of exons | 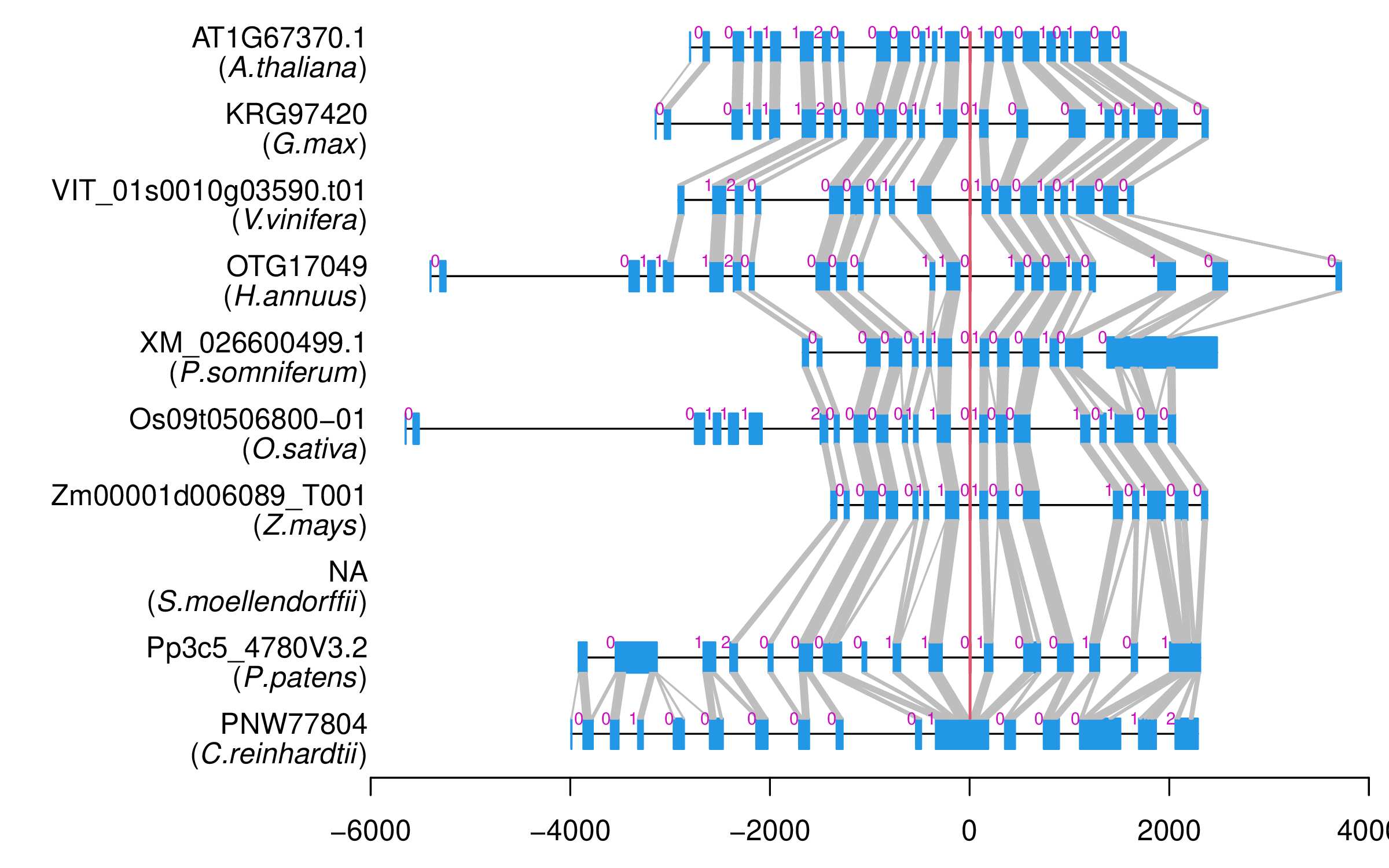 |
| Microexon DNA seq | GCTCTGACTGAAG |
| Microexon Amino Acid seq | ALTEE |
| Microexon-tag DNA Seq | AATATTGAACTGACTGATGTTCTCTCTAGCTTCCCGGACATATCAGTGGCTCTGACTGAAGAAATCATGGATAAACTTGTAAACCAAAATATACTGTCAAGAACGGGA |
| Microexon-tag Amino Acid Seq | NIELTDVLSSFPDISVALTEEIMDKLVNQNILSRTG |
| Microexon-tag spanning region | 18352537-18357856 |
| Microexon-tag prediction score | 0.9881 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | XM_026554287.1x |
| Reference Transcript ID | XM_026554287.1 |
| Gene ID | NA |
| Gene Name | NA |
| Transcript ID | XM_026554287.1 |
| Protein ID | XP_026410072.1 |
| Gene ID | LOC113305202 |
| Gene Name | NA |
| Pfam domain motif | DUF5457 |
| Motif E-value | 0.13 |
| Motif start | 37 |
| Motif end | 90 |
| Protein seq | >XP_026410072.1 MADQEDDTHDAEEDEHQLKRVRDWISSSHIDNIELTDVLSSFPDISVALTEEIMDKLVNQNILSRTGKDSYSISKMKAIV VKEEIQVNQVDEKVQRGNDEDYLYLKALYHVLPLDYVTISKLQSKLEGEANQATARELLDKMACDGYVEAKNKPRLGKRV IRSELTDKKLFEVKNVLEIKLSAMEISEPQNISTCGGLHSVGSDLTRTRERSVDTQQNGSSRIQEQLGNNTPIRGNERQG RVVYQGSISEMVWMGEDSVTVVRVIIPSVVVPVRKSAPGKLAR* |
| CDS seq | >XM_026554287.1 ATGGCGGATCAAGAAGATGATACACATGATGCAGAAGAGGACGAACATCAGTTAAAACGTGTAAGAGACTGGATTAGCTC TAGCCACATTGACAATATTGAACTGACTGATGTTCTCTCTAGCTTCCCGGACATATCAGTGGCTCTGACTGAAGAAATCA TGGATAAACTTGTAAACCAAAATATACTGTCAAGAACGGGAAAGGATTCTTACTCCATCAGCAAGATGAAGGCAATTGTC GTGAAGGAAGAGATACAAGTAAACCAAGTTGATGAGAAAGTTCAGCGAGGGAATGATGAAGACTACTTGTACTTGAAGGC TCTTTATCATGTCCTTCCACTGGATTATGTTACAATATCTAAACTTCAAAGTAAGCTGGAAGGAGAGGCAAATCAGGCAA CTGCGCGTGAACTGCTCGATAAGATGGCTTGTGATGGATATGTGGAAGCCAAGAACAAACCAAGATTAGGTAAACGTGTC ATTCGATCCGAATTAACTGACAAGAAGCTATTTGAGGTCAAGAATGTTTTGGAGATCAAGTTATCGGCGATGGAGATAAG TGAACCTCAAAACATATCAACCTGCGGTGGTCTCCACTCTGTTGGTTCTGATCTTACTAGAACTCGAGAAAGATCAGTTG ATACTCAGCAGAATGGGTCGTCTAGAATTCAGGAGCAACTTGGAAACAACACACCCATTCGTGGAAATGAGCGTCAAGGG AGAGTGGTATACCAGGGATCGATAAGCGAAATGGTATGGATGGGAGAGGATTCAGTAACTGTCGTGAGGGTGATAATACC TTCTGTAGTCGTCCCAGTCAGGAAAAGCGCTCCAGGAAAACTAGCACGGTGA |
| Microexon DNA seq | GCTCTGACTGAAG |
| Microexon Amino Acid seq | ALTEE |
| Microexon-tag DNA Seq | AATATTGAACTGACTGATGTTCTCTCTAGCTTCCCGGACATATCAGTGGCTCTGACTGAAGAAATCATGGATAAACTTGTAAACCAAAATATACTGTCAAGAACGGGA |
| Microexon-tag Amino Acid seq | NIELTDVLSSFPDISVALTEEIMDKLVNQNILSRTG |
| Transcript ID | XM_026554287.1 |
| Gene ID | Ps.48213 |
| Gene Name | NA |
| Pfam domain motif | DUF5457 |
| Motif E-value | 0.13 |
| Motif start | 37 |
| Motif end | 90 |
| Protein seq | >XM_026554287.1 MADQEDDTHDAEEDEHQLKRVRDWISSSHIDNIELTDVLSSFPDISVALTEEIMDKLVNQNILSRTGKDSYSISKMKAIV VKEEIQVNQVDEKVQRGNDEDYLYLKALYHVLPLDYVTISKLQSKLEGEANQATARELLDKMACDGYVEAKNKPRLGKRV IRSELTDKKLFEVKNVLEIKLSAMEISEPQNISTCGGLHSVGSDLTRTRERSVDTQQNGSSRIQEQLGNNTPIRGNERQG RVVYQGSISEMVWMGEDSVTVVRVIIPSVVVPVRKSAPGKLAR* |
| CDS seq | >XM_026554287.1 ATGGCGGATCAAGAAGATGATACACATGATGCAGAAGAGGACGAACATCAGTTAAAACGTGTAAGAGACTGGATTAGCTC TAGCCACATTGACAATATTGAACTGACTGATGTTCTCTCTAGCTTCCCGGACATATCAGTGGCTCTGACTGAAGAAATCA TGGATAAACTTGTAAACCAAAATATACTGTCAAGAACGGGAAAGGATTCTTACTCCATCAGCAAGATGAAGGCAATTGTC GTGAAGGAAGAGATACAAGTAAACCAAGTTGATGAGAAAGTTCAGCGAGGGAATGATGAAGACTACTTGTACTTGAAGGC TCTTTATCATGTCCTTCCACTGGATTATGTTACAATATCTAAACTTCAAAGTAAGCTGGAAGGAGAGGCAAATCAGGCAA CTGCGCGTGAACTGCTCGATAAGATGGCTTGTGATGGATATGTGGAAGCCAAGAACAAACCAAGATTAGGTAAACGTGTC ATTCGATCCGAATTAACTGACAAGAAGCTATTTGAGGTCAAGAATGTTTTGGAGATCAAGTTATCGGCGATGGAGATAAG TGAACCTCAAAACATATCAACCTGCGGTGGTCTCCACTCTGTTGGTTCTGATCTTACTAGAACTCGAGAAAGATCAGTTG ATACTCAGCAGAATGGGTCGTCTAGAATTCAGGAGCAACTTGGAAACAACACACCCATTCGTGGAAATGAGCGTCAAGGG AGAGTGGTATACCAGGGATCGATAAGCGAAATGGTATGGATGGGAGAGGATTCAGTAACTGTCGTGAGGGTGATAATACC TTCTGTAGTCGTCCCAGTCAGGAAAAGCGCTCCAGGAAAACTAGCACGGTGA |