
| Microexon ID | Vv_12:2090951-2090963:+ |
| Species | Vistis vinifera | Coordinates | 12:2090951..2090963 |
| Microexon Cluster ID | MEP31 |
| Size | 13 |
| Phase | 0 |
| Pfam Domain Motif | TPT |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,13,47 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GCTGTYATGTCTGGKTTYCGYTGGTSYATGACTCARATTCTTYTGCAGAAAGAARMHTAYGGTYTRAARAATCCAYTTACCTTGATGAGYTATGTKACYCCAGTGATG |
| Logo of Microexon-tag DNA Seq | 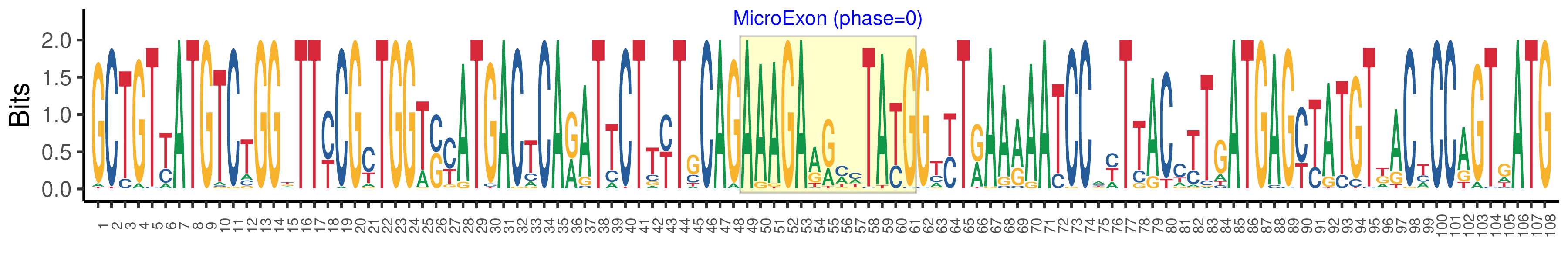 |
| Alignment of exons | 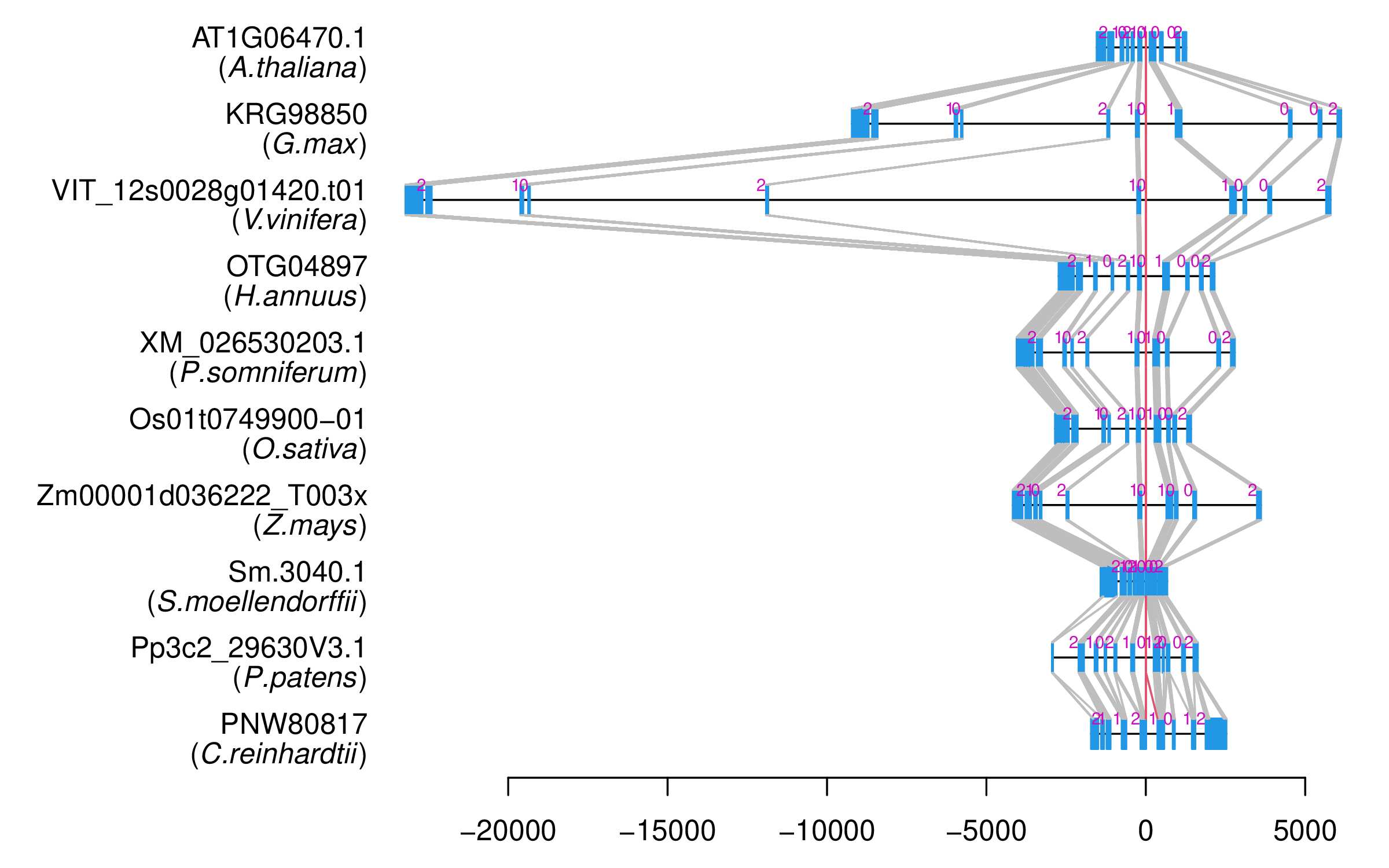 |
| Microexon DNA seq | AAAGAAGCCTATG |
| Microexon Amino Acid seq | KEAYG |
| Microexon-tag DNA Seq | GCTGTTATGTCTGGGTTCCGCTGGTCCATGACTCAGATTCTTCTCCAGAAAGAAGCCTATGGTTTGAAAAATCCACTTACATTGATGAGCTATGTGGCTCCAGTGATG |
| Microexon-tag Amino Acid Seq | AVMSGFRWSMTQILLQKEAYGLKNPLTLMSYVAPVM |
| Microexon-tag spanning region | 2090735-2093641 |
| Microexon-tag prediction score | 0.9923 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | VIT_12s0028g01420.t01x |
| Reference Transcript ID | VIT_12s0028g01420.t01 |
| Gene ID | VIT_12s0028g01420 |
| Gene Name | NA |
| Transcript ID | VIT_12s0028g01420.t01 |
| Protein ID | VIT_12s0028g01420.t01 |
| Gene ID | VIT_12s0028g01420 |
| Gene Name | NA |
| Pfam domain motif | TPT |
| Motif E-value | 1.4e-28 |
| Motif start | 162 |
| Motif end | 461 |
| Protein seq | >VIT_12s0028g01420.t01 MVESDLEIEEVSYKGINYTQTGNQGPVLRREPSFSRWCDEDGIICSGCQLDNAEAGSEDSDFELPLLQQGELENSVSDKE RYQYSKFEKRTMQLNGGNTMDGSSIHVRGNGNEKYVPFDIEDKPEREIDFSNIGVDGSNSNGNHEMSSPNSNNSIDVVSL LKTLFFILVWYTFSTILTLYNKTLLGDDLGRFPAPLLMNTVHFLMQAILSKAITCFWSQRFQPSVTMSWRDYFVRVVPTA LGTALDINLSNASLVFISVTFATMCKSASPIFLLIFAFAFRLESPSIKLLGIMMIISIGILLTVAKETEFEIWGFIFVML AAVMSGFRWSMTQILLQKEAYGLKNPLTLMSYVAPVMTVATALLSLLMDPWYEFKTNNYFNSSWHVIRSCLLMLIGGTLA FFMVLTEYILVSVTSAVTVTIAGVVKEAVTILVAVFYFHDQFTWLKGVGLSTIMVGVSLFNWYKYLKLQTGHSNEVDMGD TAGDIAGSLKSNINARYVILEEVDEQEDST* |
| CDS seq | >VIT_12s0028g01420.t01 ATGGTAGAAAGTGATTTAGAAATAGAAGAAGTTTCATACAAGGGGATTAACTATACTCAGACTGGAAACCAAGGGCCAGT TCTGCGCAGGGAGCCCTCATTCTCACGCTGGTGTGATGAAGATGGGATAATTTGTTCAGGTTGTCAATTAGACAATGCAG AAGCAGGTTCAGAAGACTCTGATTTTGAATTGCCTTTGCTTCAACAGGGTGAACTAGAAAATAGTGTTTCAGACAAGGAG AGGTACCAATATAGTAAGTTTGAGAAGAGAACTATGCAATTGAATGGTGGTAACACAATGGATGGTTCTTCTATTCATGT TAGGGGAAATGGAAATGAGAAGTATGTGCCCTTTGATATTGAAGATAAACCTGAAAGAGAGATAGACTTCTCAAATATTG GTGTGGATGGCTCAAATTCTAACGGAAACCATGAAATGTCGTCGCCAAATTCCAATAACTCTATTGATGTTGTTAGTTTG CTGAAGACATTGTTTTTTATTCTTGTTTGGTATACTTTCAGCACAATCTTGACACTGTACAATAAAACACTGCTAGGAGA TGATTTGGGAAGGTTCCCTGCTCCTTTATTGATGAATACTGTGCACTTTCTAATGCAAGCAATTTTATCAAAAGCCATCA CATGCTTTTGGTCTCAAAGATTTCAACCTAGCGTGACTATGTCTTGGAGAGACTACTTTGTGAGGGTTGTACCAACAGCT CTTGGAACAGCATTGGATATTAACTTGAGCAATGCATCCCTTGTTTTCATCTCTGTTACATTTGCAACTATGTGTAAATC TGCTTCTCCCATATTTCTCCTTATATTTGCTTTTGCATTCAGGTTGGAATCTCCCAGCATTAAACTTCTAGGTATTATGA TGATTATCTCCATTGGAATTTTATTAACAGTTGCAAAAGAGACAGAGTTTGAGATTTGGGGTTTCATTTTTGTTATGCTT GCTGCTGTTATGTCTGGGTTCCGCTGGTCCATGACTCAGATTCTTCTCCAGAAAGAAGCCTATGGTTTGAAAAATCCACT TACATTGATGAGCTATGTGGCTCCAGTGATGACTGTGGCTACCGCTCTACTTTCTCTTCTAATGGACCCGTGGTATGAAT TTAAAACGAACAATTACTTCAATAGTTCATGGCATGTCATACGGAGCTGCTTGCTGATGCTAATTGGTGGAACTCTGGCC TTCTTTATGGTTTTGACAGAATACATTCTTGTCTCAGTAACTAGTGCTGTAACAGTGACTATAGCAGGGGTTGTCAAGGA GGCTGTCACTATTTTGGTTGCAGTCTTTTACTTCCATGATCAGTTTACCTGGTTGAAGGGGGTTGGTCTATCCACAATCA TGGTTGGTGTTAGTTTATTCAACTGGTACAAATATCTAAAGCTACAGACGGGCCACAGCAATGAAGTTGACATGGGTGAT ACTGCTGGTGATATTGCTGGCTCACTCAAATCAAATATTAATGCAAGATATGTTATTCTTGAGGAGGTGGATGAGCAAGA GGATAGCACTTGA |
| Microexon DNA seq | AAAGAAGCCTATG |
| Microexon Amino Acid seq | KEAYG |
| Microexon-tag DNA Seq | GCTGTTATGTCTGGGTTCCGCTGGTCCATGACTCAGATTCTTCTCCAGAAAGAAGCCTATGGTTTGAAAAATCCACTTACATTGATGAGCTATGTGGCTCCAGTGATG |
| Microexon-tag Amino Acid seq | AVMSGFRWSMTQILLQKEAYGLKNPLTLMSYVAPVM |
| Transcript ID | Vv.4755.1 |
| Gene ID | Vv.4755 |
| Gene Name | NA |
| Pfam domain motif | TPT |
| Motif E-value | 1.4e-28 |
| Motif start | 162 |
| Motif end | 461 |
| Protein seq | >Vv.4755.1 MVESDLEIEEVSYKGINYTQTGNQGPVLRREPSFSRWCDEDGIICSGCQLDNAEAGSEDSDFELPLLQQGELENSVSDKE RYQYSKFEKRTMQLNGGNTMDGSSIHVRGNGNEKYVPFDIEDKPEREIDFSNIGVDGSNSNGNHEMSSPNSNNSIDVVSL LKTLFFILVWYTFSTILTLYNKTLLGDDLGRFPAPLLMNTVHFLMQAILSKAITCFWSQRFQPSVTMSWRDYFVRVVPTA LGTALDINLSNASLVFISVTFATMCKSASPIFLLIFAFAFRLESPSIKLLGIMMIISIGILLTVAKETEFEIWGFIFVML AAVMSGFRWSMTQILLQKEAYGLKNPLTLMSYVAPVMTVATALLSLLMDPWYEFKTNNYFNSSWHVIRSCLLMLIGGTLA FFMVLTEYILVSVTSAVTVTIAGVVKEAVTILVAVFYFHDQFTWLKGVGLSTIMVGVSLFNWYKYLKLQTGHSNEVDMGD TAGDIAGDIAGSLKSNINARYVILEEVDEQEDST* |
| CDS seq | >Vv.4755.1 ATGGTAGAAAGTGATTTAGAAATAGAAGAAGTTTCATACAAGGGGATTAACTATACTCAGACTGGAAACCAAGGGCCAGT TCTGCGCAGGGAGCCCTCATTCTCACGCTGGTGTGATGAAGATGGGATAATTTGTTCAGGTTGTCAATTAGACAATGCAG AAGCAGGTTCAGAAGACTCTGATTTTGAATTGCCTTTGCTTCAACAGGGTGAACTAGAAAATAGTGTTTCAGACAAGGAG AGGTACCAATATAGTAAGTTTGAGAAGAGAACTATGCAATTGAATGGTGGTAACACAATGGATGGTTCTTCTATTCATGT TAGGGGAAATGGAAATGAGAAGTATGTGCCCTTTGATATTGAAGATAAACCTGAAAGAGAGATAGACTTCTCAAATATTG GTGTGGATGGCTCAAATTCTAACGGAAACCATGAAATGTCGTCGCCAAATTCCAATAACTCTATTGATGTTGTTAGTTTG CTGAAGACATTGTTTTTTATTCTTGTTTGGTATACTTTCAGCACAATCTTGACACTGTACAATAAAACACTGCTAGGAGA TGATTTGGGAAGGTTCCCTGCTCCTTTATTGATGAATACTGTGCACTTTCTAATGCAAGCAATTTTATCAAAAGCCATCA CATGCTTTTGGTCTCAAAGATTTCAACCTAGCGTGACTATGTCTTGGAGAGACTACTTTGTGAGGGTTGTACCAACAGCT CTTGGAACAGCATTGGATATTAACTTGAGCAATGCATCCCTTGTTTTCATCTCTGTTACATTTGCAACTATGTGTAAATC TGCTTCTCCCATATTTCTCCTTATATTTGCTTTTGCATTCAGGTTGGAATCTCCCAGCATTAAACTTCTAGGTATTATGA TGATTATCTCCATTGGAATTTTATTAACAGTTGCAAAAGAGACAGAGTTTGAGATTTGGGGTTTCATTTTTGTTATGCTT GCTGCTGTTATGTCTGGGTTCCGCTGGTCCATGACTCAGATTCTTCTCCAGAAAGAAGCCTATGGTTTGAAAAATCCACT TACATTGATGAGCTATGTGGCTCCAGTGATGACTGTGGCTACCGCTCTACTTTCTCTTCTAATGGACCCGTGGTATGAAT TTAAAACGAACAATTACTTCAATAGTTCATGGCATGTCATACGGAGCTGCTTGCTGATGCTAATTGGTGGAACTCTGGCC TTCTTTATGGTTTTGACAGAATACATTCTTGTCTCAGTAACTAGTGCTGTAACAGTGACTATAGCAGGGGTTGTCAAGGA GGCTGTCACTATTTTGGTTGCAGTCTTTTACTTCCATGATCAGTTTACCTGGTTGAAGGGGGTTGGTCTATCCACAATCA TGGTTGGTGTTAGTTTATTCAACTGGTACAAATATCTAAAGCTACAGACGGGCCACAGCAATGAAGTTGACATGGGTGAT ACTGCTGGTGATATTGCTGGCGATATTGCTGGCTCACTCAAATCAAATATTAATGCAAGATATGTTATTCTTGAGGAGGT GGATGAGCAAGAGGATAGCACTTGA |