
| Microexon ID | Ta_3D:399297110-399297122:+ |
| Species | Triticum Aestivum | Coordinates | 3D:399297110..399297122 |
| Microexon Cluster ID | MEP31 |
| Size | 13 |
| Phase | 0 |
| Pfam Domain Motif | TPT |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,13,47 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GCTGTYATGTCTGGKTTYCGYTGGTSYATGACTCARATTCTTYTGCAGAAAGAARMHTAYGGTYTRAARAATCCAYTTACCTTGATGAGYTATGTKACYCCAGTGATG |
| Logo of Microexon-tag DNA Seq | 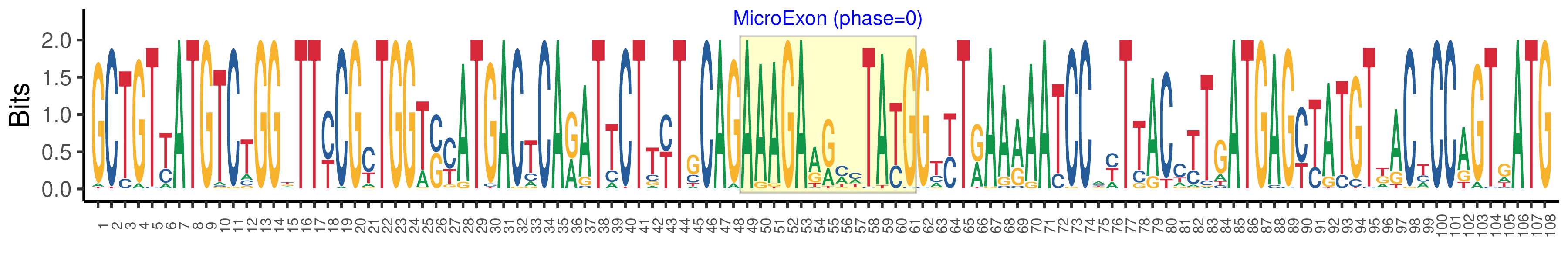 |
| Alignment of exons | 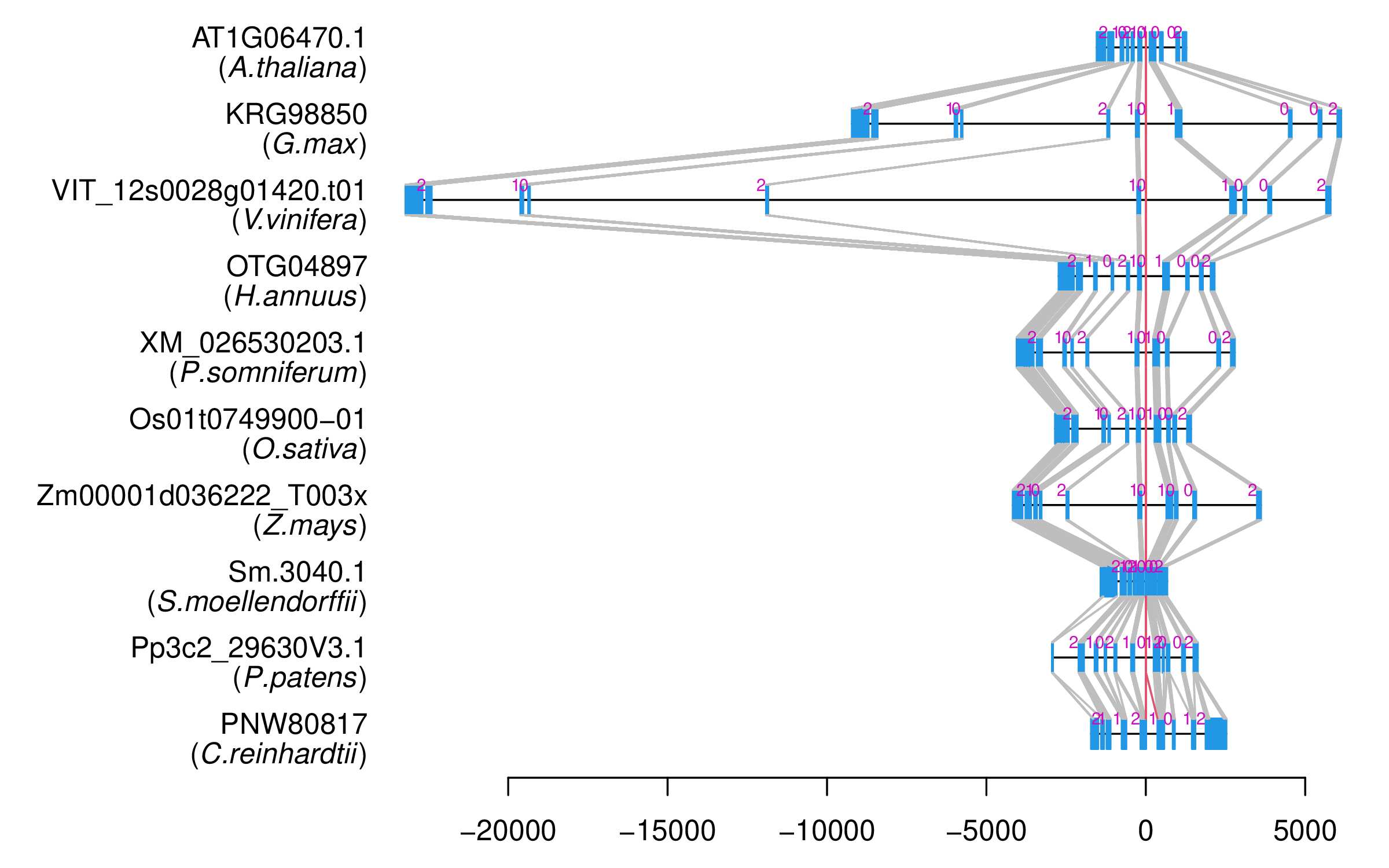 |
| Microexon DNA seq | AAAGAGGAATATG |
| Microexon Amino Acid seq | KEEYG |
| Microexon-tag DNA Seq | GCTGTCATGTCTGGTTTCCGCTGGTGCATGACACAGATTCTCCTCCAGAAAGAGGAATATGGATTAAAGAACCCCTTCACCTTAATGAGTTATGTTACCCCGGTGATG |
| Microexon-tag Amino Acid Seq | AVMSGFRWCMTQILLQKEEYGLKNPFTLMSYVTPVM |
| Microexon-tag spanning region | 399296901-399297488 |
| Microexon-tag prediction score | 0.9688 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | TraesCS3D02G289300.3x |
| Reference Transcript ID | TraesCS3D02G289300.3 |
| Gene ID | TraesCS3D02G289300 |
| Gene Name | NA |
| Transcript ID | TraesCS3D02G289300.3 |
| Protein ID | |
| Gene ID | TraesCS3D02G289300 |
| Gene Name | |
| Pfam domain motif | TPT |
| Motif E-value | 1.70E-30 |
| Motif start | 121 |
| Motif end | 421 |
| Protein seq | >TraesCS3D02G289300.3 MGDCAVENGHAHPKEEDAPEVPVEPAEGAAEPQVVAGGQRKQRGIRREPSFSRWCKDPSAAPNAPAGAASPASAPSDDDS EEFELPLLPPAAAAHRSPMDIEAGAAAGSDSVPVSPWLIAKIIFLIASWYTLSTCLTLYNKEMLGKRMWKFPAPFLMNTV HFTMQAVASRVIVWFQQRGMEAERNPMSWKDYFLRVVPTALATALDINLTNISFVFITVTFATMCKSGAPIFILLFAFLF RLEKPSFNILGIMLIVSVGVLLTVAKETQFNLWGFIFIMLAAVMSGFRWCMTQILLQKEEYGLKNPFTLMSYVTPVMAIT TAIISIAMDPWHEVRASHFFDSPAHILRSILLMLLGGALAFFMVLTEYVLVSVTSAVTVTIAGIVKEAVTILVAVLFFND PFTWLKGFGLATIIFGVSLFNLYKYHRFKKDRHSKHVDPNSHSSNGASKYVIIDDDIEDLDDTG* |
| CDS seq | >TraesCS3D02G289300.3 ATGGGGGACTGCGCCGTGGAGAACGGCCACGCCCACCCGAAGGAAGAGGACGCGCCGGAAGTGCCCGTCGAGCCCGCGGA GGGCGCCGCGGAGCCGCAGGTGGTGGCTGGCGGGCAGAGGAAGCAGAGAGGGATACGCAGGGAGCCGTCGTTCTCGCGGT GGTGCAAGGACCCCTCCGCGGCGCCCAATGCCCCCGCCGGCGCCGCTTCGCCGGCCTCTGCACCGAGCGACGATGATTCG GAGGAGTTCGAGCTGCCGCTTCTGCCGCCCGCCGCCGCCGCCCATCGCTCGCCGATGGATATCGAGGCAGGAGCGGCGGC GGGATCTGACAGCGTCCCGGTCTCGCCGTGGCTGATCGCCAAGATTATATTTCTGATAGCGAGTTGGTACACGCTGAGCA CCTGCTTGACGCTGTATAACAAGGAGATGCTCGGGAAGCGCATGTGGAAGTTCCCTGCCCCGTTCCTGATGAACACGGTG CATTTCACAATGCAGGCTGTTGCGTCGAGGGTGATCGTGTGGTTCCAGCAACGGGGCATGGAGGCAGAGCGTAATCCAAT GTCATGGAAAGATTACTTTCTGCGAGTTGTTCCAACAGCTTTGGCTACTGCTCTTGATATAAATTTGACCAACATTTCGT TTGTTTTCATTACTGTGACATTCGCTACAATGTGTAAATCTGGGGCTCCAATTTTCATTCTTCTCTTTGCTTTTCTATTC AGGCTAGAGAAGCCGAGCTTTAACATTCTTGGAATCATGCTGATTGTTTCTGTTGGAGTTCTACTGACAGTTGCTAAAGA GACACAATTTAATCTTTGGGGATTTATATTTATTATGCTTGCTGCTGTCATGTCTGGTTTCCGCTGGTGCATGACACAGA TTCTCCTCCAGAAAGAGGAATATGGATTAAAGAACCCCTTCACCTTAATGAGTTATGTTACCCCGGTGATGGCAATAACA ACTGCGATTATTTCCATTGCCATGGATCCATGGCATGAAGTCAGAGCAAGTCACTTCTTTGATAGTCCAGCTCACATATT AAGAAGCATCTTATTGATGCTTCTAGGTGGTGCTTTGGCATTCTTCATGGTTTTGACAGAATATGTTCTGGTTTCTGTTA CAAGTGCTGTAACAGTGACCATAGCTGGGATTGTCAAGGAAGCTGTCACAATTTTGGTTGCTGTGCTTTTCTTCAATGAT CCATTTACCTGGTTAAAGGGATTTGGACTTGCAACAATTATATTTGGTGTCAGCCTTTTCAATCTGTACAAGTATCATAG GTTCAAGAAAGACCGCCACAGCAAACATGTTGACCCAAATTCCCATTCCTCCAATGGTGCCTCAAAATATGTTATCATTG ATGATGATATTGAAGATCTGGATGATACAGGCTGA |
Ta_3D:399297110-399297122:+ does not have available information here.