
| Microexon ID | Sb_3:62828864-62828876:+ |
| Species | Sorghum Bicolor | Coordinates | 3:62828864..62828876 |
| Microexon Cluster ID | MEP31 |
| Size | 13 |
| Phase | 0 |
| Pfam Domain Motif | TPT |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,13,47 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GCTGTYATGTCTGGKTTYCGYTGGTSYATGACTCARATTCTTYTGCAGAAAGAARMHTAYGGTYTRAARAATCCAYTTACCTTGATGAGYTATGTKACYCCAGTGATG |
| Logo of Microexon-tag DNA Seq | 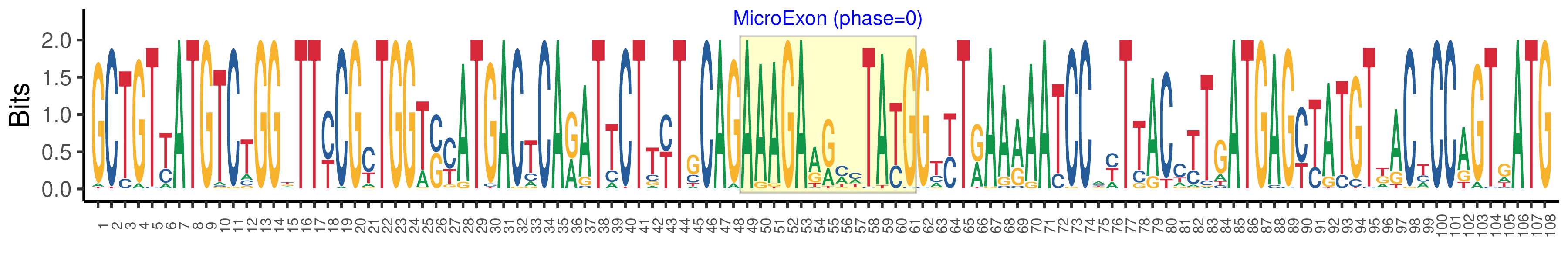 |
| Alignment of exons |  |
| Microexon DNA seq | AAAGAGGAATACG |
| Microexon Amino Acid seq | KEEYG |
| Microexon-tag DNA Seq | GCTGTTATGTCTGGTTTCCGCTGGTCCATGACTCAGATTCTTCTCCAGAAAGAGGAATACGGATTAAAGAATCCTTTTACCTTGATGAGCCATGTTACCCCAGTAATG |
| Microexon-tag Amino Acid Seq | AVMSGFRWSMTQILLQKEEYGLKNPFTLMSHVTPVM |
| Microexon-tag spanning region | 62828654-62829935 |
| Microexon-tag prediction score | 0.9827 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KXG33369x |
| Reference Transcript ID | KXG33369 |
| Gene ID | SORBI_3003G296200 |
| Gene Name | NA |
| Transcript ID | KXG33369 |
| Protein ID | |
| Gene ID | SORBI_3003G296200 |
| Gene Name | |
| Pfam domain motif | TPT |
| Motif E-value | 8.70E-31 |
| Motif start | 126 |
| Motif end | 425 |
| Protein seq | >KXG33369 MGDCVVEDGHGHHQSEAVEGAVPLTVAVELDERSGESREEGGGQRKKVGGIRREPSFSRWCRDPSDAAASNTAAAAATSD SDDSEEFELPLLPSSSGGGSSPMDIEAGATARSDDLPISPRLLAKVIGLIACWYTLSTCLTLYNKEMLGKHMWKFPAPFL MNTVHFTLQAVASRAIVWFQQRGLEGGPSKMSWKDYCLRVVPTALATALDINLSNISLVFITVTFATMCKSASPIFILLF AFMFRLEKPSFSLLGIMLVVSFGVLLTVAKETEFNLWGFIFIMLAAVMSGFRWSMTQILLQKEEYGLKNPFTLMSHVTPV MAIVTAIISIVMDPWHDFRASHFFDSSAHIIRSSLLLLLGGALAFFMVLTEYVLVSVTSAVTVTVAGIVKEAVTILVAVL FFHDPFTWLKALGLAIIIFGVSLFNIYKYKRFKKGHHNEDTGTNIQSSNGTSKYVILDDDTEAQDDTG* |
| CDS seq | >KXG33369 ATGGGGGACTGCGTTGTCGAGGACGGCCACGGGCACCACCAATCGGAGGCGGTGGAGGGCGCGGTTCCGTTGACGGTAGC CGTGGAGCTCGATGAGCGGTCGGGAGAGTCGCGGGAGGAGGGTGGCGGGCAGCGGAAGAAGGTTGGTGGGATCCGAAGGG AGCCGTCGTTCTCGCGGTGGTGCAGGGACCCCTCTGACGCCGCCGCCTCCAATACCGCCGCTGCTGCGGCGACCAGCGAC AGCGACGACTCAGAGGAGTTCGAGCTGCCGCTCCTGCCATCCTCCTCGGGCGGCGGCAGCTCACCGATGGACATTGAGGC GGGGGCGACGGCGAGATCTGATGATCTGCCGATCTCGCCGAGGTTGCTTGCGAAGGTCATCGGGTTGATAGCGTGTTGGT ACACGCTTAGCACATGCTTGACGCTGTACAACAAGGAGATGCTGGGGAAGCACATGTGGAAGTTCCCGGCGCCATTCCTG ATGAACACAGTGCATTTCACGTTGCAGGCTGTGGCGTCCAGGGCCATCGTGTGGTTCCAGCAACGCGGCCTGGAGGGAGG GCCAAGTAAAATGTCGTGGAAGGATTACTGTTTACGAGTTGTCCCAACAGCTTTGGCTACTGCTCTTGATATAAATCTGA GCAACATTTCACTTGTTTTCATTACTGTGACATTTGCTACAATGTGCAAATCGGCTTCTCCAATTTTCATTCTTCTGTTT GCTTTTATGTTCAGGCTTGAGAAACCGAGCTTTAGCCTTCTTGGAATCATGCTGGTTGTTTCATTTGGAGTTCTACTAAC AGTTGCTAAAGAGACAGAATTTAATCTGTGGGGATTTATATTTATTATGCTTGCTGCTGTTATGTCTGGTTTCCGCTGGT CCATGACTCAGATTCTTCTCCAGAAAGAGGAATACGGATTAAAGAATCCTTTTACCTTGATGAGCCATGTTACCCCAGTA ATGGCAATAGTGACAGCAATTATTTCCATTGTGATGGATCCATGGCACGACTTCAGAGCAAGCCACTTTTTTGATAGTTC TGCTCACATAATAAGAAGCAGCTTGTTGCTGCTTCTAGGTGGCGCTTTGGCCTTTTTCATGGTTTTGACAGAATATGTCC TTGTTTCTGTGACTAGTGCTGTAACAGTTACTGTAGCAGGAATTGTGAAGGAAGCTGTCACTATTCTGGTTGCTGTGCTA TTCTTTCATGATCCATTTACATGGTTAAAGGCGCTTGGGCTGGCAATAATAATATTTGGTGTCAGTCTATTCAATATTTA CAAGTATAAAAGGTTCAAAAAAGGACATCACAATGAAGATACTGGGACAAATATCCAATCTTCCAATGGGACTTCAAAGT ACGTTATCCTTGACGATGATACTGAAGCCCAAGATGATACAGGCTGA |
Sb_3:62828864-62828876:+ does not have available information here.