
| Microexon ID | Sb_10:3293646-3293658:+ |
| Species | Sorghum Bicolor | Coordinates | 10:3293646..3293658 |
| Microexon Cluster ID | MEP31 |
| Size | 13 |
| Phase | 0 |
| Pfam Domain Motif | TPT |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,13,47 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GCTGTYATGTCTGGKTTYCGYTGGTSYATGACTCARATTCTTYTGCAGAAAGAARMHTAYGGTYTRAARAATCCAYTTACCTTGATGAGYTATGTKACYCCAGTGATG |
| Logo of Microexon-tag DNA Seq | 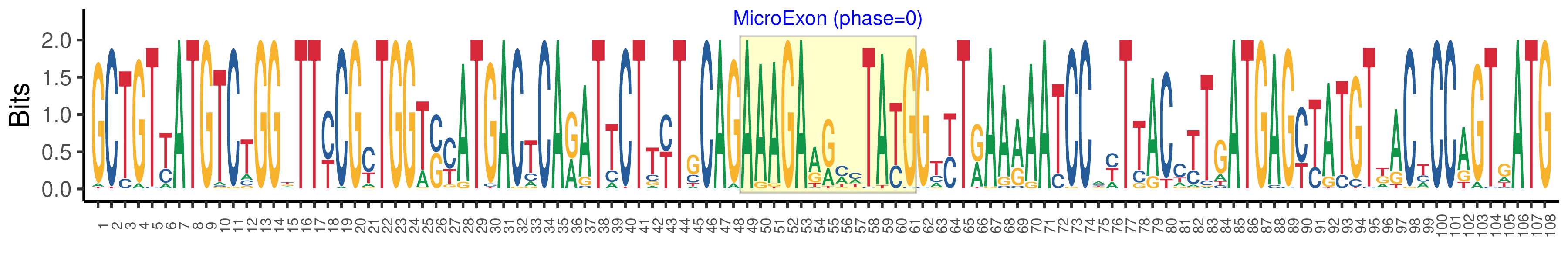 |
| Alignment of exons |  |
| Microexon DNA seq | AAAGACACTTACG |
| Microexon Amino Acid seq | KDTYG |
| Microexon-tag DNA Seq | GCTGTTATGTCGGGATTCCGTTGGTCCATGACCCAGATTCTGTTGCAGAAAGACACTTACGGCCTGAAGAATCCAATTACCTTGATGAGCCATGTTACACCAGTGATG |
| Microexon-tag Amino Acid Seq | AVMSGFRWSMTQILLQKDTYGLKNPITLMSHVTPVM |
| Microexon-tag spanning region | 3293463-3294317 |
| Microexon-tag prediction score | 0.9703 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KXG19328x |
| Reference Transcript ID | KXG19328 |
| Gene ID | SORBI_3010G042600 |
| Gene Name | NA |
| Transcript ID | KXG19328 |
| Protein ID | |
| Gene ID | SORBI_3010G042600 |
| Gene Name | |
| Pfam domain motif | TPT |
| Motif E-value | 2.00E-26 |
| Motif start | 91 |
| Motif end | 390 |
| Protein seq | >KXG19328 MHPKPEGGDADGAAAAEVGSPPPGYFRQRSMHAAAAADPEAAAARRAFDVENPPCSAGGAGGGLRPSESVTKLESLERAE RAALAPAVVLRTGFYILVWYAFSTCLTLYNKTLLGDKLGKFPAPLLMNTVHFALQAGLSKLIIFFQSKGPEAAIEMGWKD YLMRVVPTALGTALDINLSNASLVFISVTFATMCKSASPIFLLLFAFAFRLENPSIKLLGIIVVISTGVLLTVSKETEFD FWGFIFVTLAAVMSGFRWSMTQILLQKDTYGLKNPITLMSHVTPVMAIATMILSLLLDPWSEFQNNSYFDNPWHVVRSCL LMLIGGSLAFFMVLTEYILISATSAITVTIAGVVKEAVTILVAVFYFHDEFTWLKGFGLFTIMVGVSLFNWYKYEKFKRG QTNEDEVNSPPFTGDVKYIILDDLEYQDEFEEEDT* |
| CDS seq | >KXG19328 ATGCATCCCAAGCCCGAGGGCGGCGACGCCGACGGGGCCGCCGCCGCGGAGGTGGGATCCCCGCCGCCGGGCTACTTCCG GCAGCGGAGCATGCACGCCGCCGCCGCCGCGGATCCGGAGGCGGCGGCGGCGCGCCGGGCGTTCGACGTCGAGAACCCGC CTTGCTCCGCCGGCGGAGCGGGCGGGGGGCTCCGGCCCAGCGAGTCCGTCACCAAGCTGGAGTCGCTGGAGCGCGCGGAG CGCGCCGCGCTGGCGCCCGCCGTCGTGCTCAGGACCGGGTTCTACATCCTGGTGTGGTACGCGTTCAGCACCTGCCTCAC GCTATATAACAAGACGCTGCTGGGGGATAAGCTCGGCAAGTTCCCGGCGCCGCTGCTGATGAACACCGTGCACTTCGCAC TGCAGGCGGGGCTCTCCAAGCTCATAATCTTCTTCCAGTCCAAGGGGCCCGAGGCCGCCATCGAGATGGGCTGGAAGGAC TACTTGATGAGAGTCGTGCCAACTGCTCTTGGGACGGCCTTGGACATAAACCTGAGCAATGCGTCTCTGGTGTTCATCTC GGTGACGTTTGCTACTATGTGTAAATCAGCCTCCCCGATATTCCTGTTGCTCTTTGCGTTTGCATTTAGGTTGGAGAATC CTAGCATCAAGCTTCTAGGAATTATAGTTGTTATTTCTACTGGAGTTCTGTTGACAGTTTCCAAGGAGACAGAGTTTGAC TTCTGGGGCTTCATTTTTGTGACGCTTGCTGCTGTTATGTCGGGATTCCGTTGGTCCATGACCCAGATTCTGTTGCAGAA AGACACTTACGGCCTGAAGAATCCAATTACCTTGATGAGCCATGTTACACCAGTGATGGCCATAGCCACTATGATACTAT CTCTATTACTGGATCCTTGGAGTGAATTTCAAAATAATTCATATTTCGATAACCCTTGGCATGTTGTGCGCAGTTGCTTA CTCATGCTTATCGGAGGATCCTTAGCTTTTTTCATGGTGTTAACAGAGTATATACTTATTTCAGCAACAAGTGCCATAAC CGTGACGATAGCTGGGGTAGTAAAAGAGGCTGTAACCATTTTGGTTGCAGTATTTTATTTCCATGATGAATTTACTTGGT TGAAAGGGTTTGGTCTGTTCACCATCATGGTTGGTGTTAGCTTGTTCAATTGGTACAAGTATGAGAAATTCAAAAGGGGT CAGACAAATGAGGATGAAGTCAACTCTCCTCCATTTACTGGTGACGTCAAGTACATTATTCTAGATGACTTGGAATATCA GGATGAATTTGAGGAGGAAGATACATAA |
Sb_10:3293646-3293658:+ does not have available information here.