
| Microexon ID | Ps_NC_039358.1:24721338-24721350:- |
| Species | Papaver somniferum | Coordinates | NC_039358.1:24721338..24721350 |
| Microexon Cluster ID | MEP31 |
| Size | 13 |
| Phase | 0 |
| Pfam Domain Motif | TPT |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,13,47 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | GCTGTYATGTCTGGKTTYCGYTGGTSYATGACTCARATTCTTYTGCAGAAAGAARMHTAYGGTYTRAARAATCCAYTTACCTTGATGAGYTATGTKACYCCAGTGATG |
| Logo of Microexon-tag DNA Seq | 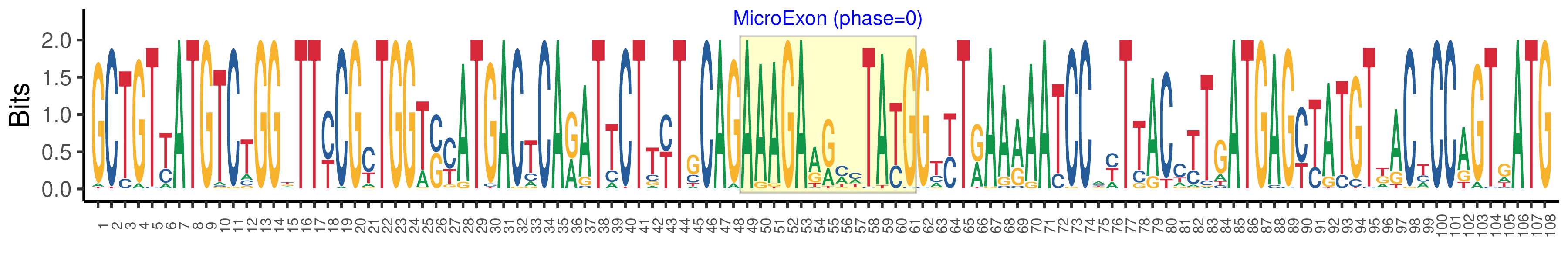 |
| Alignment of exons | 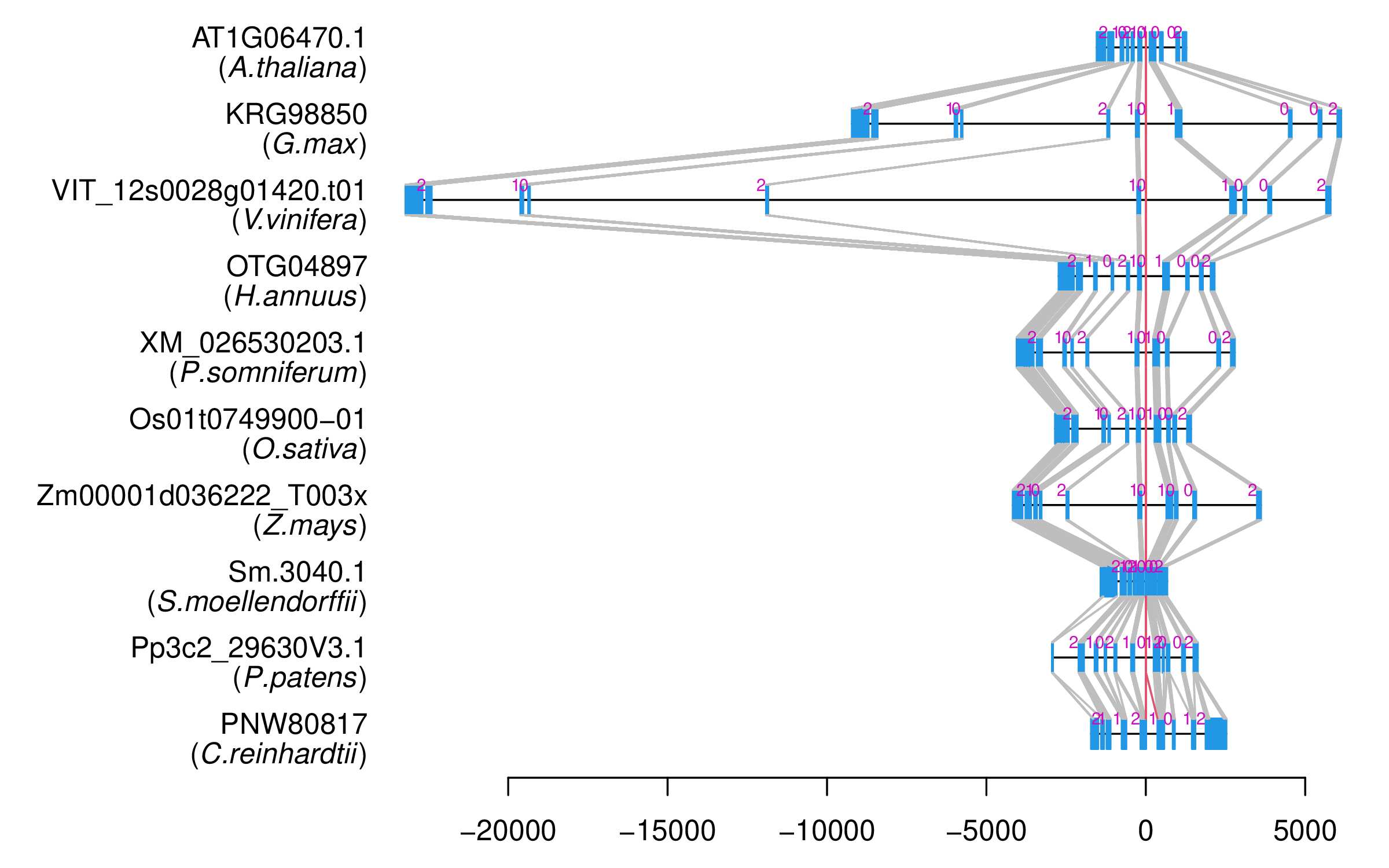 |
| Microexon DNA seq | AAAGAAACTTATG |
| Microexon Amino Acid seq | KETYG |
| Microexon-tag DNA Seq | GCTGTCATGTCTGGGTTTCGCTGGTCCATGACTCAGATTCTTTTGCAGAAAGAAACTTATGGTCTAAAGAACCCACTTACTCTGATGAGCTATGTTACTCCAGTTATG |
| Microexon-tag Amino Acid Seq | AVMSGFRWSMTQILLQKETYGLKNPLTLMSYVTPVM |
| Microexon-tag spanning region | 24721069-24721621 |
| Microexon-tag prediction score | 0.9793 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | XM_026530203.1x |
| Reference Transcript ID | XM_026530203.1 |
| Gene ID | NA |
| Gene Name | NA |
| Transcript ID | XM_026530203.1 |
| Protein ID | XP_026385988.1 |
| Gene ID | LOC113281462 |
| Gene Name | NA |
| Pfam domain motif | TPT |
| Motif E-value | 1.2e-29 |
| Motif start | 160 |
| Motif end | 459 |
| Protein seq | >XP_026385988.1 MVDSNLEVEKDDAIELQDIQKESDRPVLRREPSFSRWFDEDGVIHFDRPLEHADETVEDSDFELPFLQTEGQESKLLSKD RLSNPRNMHLNGNHTMPMVRTHNHVDENTNGLNYVPFDIENESIRELPVSDFDAHDSSSRGSHKTTLDTQNSLDMAIVLK TLFFILVWYTFSTCLTIYNKTLLGEKLGKFPAPMLMNTIHFGMQAVLSNFITWFWSNRFQPTVTMSWKDYFMRVVPTALG TALDINLSNASLVFISITFATMCKSASPIFLLLFAFAFKLESPSVKLLGIISVISVGILLTVAKETEFQFWGFVFVMLAA VMSGFRWSMTQILLQKETYGLKNPLTLMSYVTPVMAVATAILSLAMDPWDEFRKNIYFDSSWHVFRSALLMFLGGTLAFF MVLTEYILVSATSAVTVTIAGVVKEVVTILVAVFYFHDKFTWLKGVGLFIIIFGVSLFNWYKYQKHQERQTSVVDESGLS PKNVAAKYVILEEMEDHDVDT* |
| CDS seq | >XM_026530203.1 ATGGTAGACAGTAATTTGGAGGTGGAAAAGGATGATGCAATTGAGCTTCAAGATATTCAGAAAGAAAGTGACAGACCGGT TCTTCGAAGGGAACCATCTTTCTCACGCTGGTTTGACGAGGACGGCGTCATTCATTTCGATCGTCCTTTGGAGCATGCTG ATGAAACTGTAGAAGACTCTGATTTTGAGTTGCCGTTTCTTCAAACTGAAGGACAAGAGAGCAAATTATTGAGCAAGGAC CGACTGTCTAATCCGAGAAACATGCACCTGAATGGGAATCACACCATGCCCATGGTTAGGACTCATAATCATGTGGATGA AAATACAAATGGACTGAACTATGTGCCATTTGACATTGAAAATGAGTCTATTCGAGAATTGCCTGTTTCAGATTTTGATG CCCATGACTCCAGTTCAAGAGGAAGCCATAAAACGACACTGGATACACAGAATTCTCTTGATATGGCCATTGTGCTCAAG ACATTGTTTTTCATACTCGTATGGTACACTTTCAGTACATGTTTGACAATTTATAACAAAACATTATTAGGAGAAAAGTT GGGTAAATTCCCGGCTCCCATGTTGATGAATACTATCCACTTTGGGATGCAAGCTGTTCTATCGAATTTTATTACATGGT TTTGGTCTAACAGATTTCAACCCACAGTAACAATGTCATGGAAGGATTACTTCATGAGAGTTGTTCCCACGGCTCTTGGA ACTGCACTGGACATAAACCTGAGCAATGCCTCTCTTGTGTTCATCTCTATCACATTTGCCACCATGTGTAAATCAGCATC TCCTATATTTCTACTTCTATTTGCTTTTGCATTCAAGTTGGAGTCTCCAAGTGTTAAACTTCTTGGCATCATATCAGTCA TCTCTGTTGGAATTTTGTTAACAGTTGCAAAGGAAACAGAGTTTCAATTTTGGGGATTCGTATTTGTTATGCTTGCTGCT GTCATGTCTGGGTTTCGCTGGTCCATGACTCAGATTCTTTTGCAGAAAGAAACTTATGGTCTAAAGAACCCACTTACTCT GATGAGCTATGTTACTCCAGTTATGGCTGTGGCAACTGCTATACTCTCTCTTGCTATGGATCCATGGGATGAATTCAGAA AGAATATTTACTTTGATAGTTCATGGCATGTCTTTCGAAGTGCATTGTTGATGTTTCTTGGCGGAACGCTAGCCTTCTTT ATGGTTTTAACAGAATACATTCTTGTCTCAGCAACCAGTGCGGTAACTGTAACAATTGCAGGAGTCGTCAAGGAAGTTGT CACTATTTTGGTTGCGGTGTTTTACTTCCATGACAAATTTACCTGGTTGAAGGGAGTTGGGCTCTTCATAATAATCTTTG GTGTCAGTTTATTCAACTGGTACAAATATCAAAAGCATCAAGAACGCCAAACTAGTGTAGTTGATGAATCTGGGTTATCA CCCAAAAATGTAGCTGCGAAGTATGTTATCCTTGAGGAGATGGAGGATCACGATGTCGATACTTGA |
| Microexon DNA seq | AAAGAAACTTATG |
| Microexon Amino Acid seq | KETYG |
| Microexon-tag DNA Seq | GCTGTCATGTCTGGGTTTCGCTGGTCCATGACTCAGATTCTTTTGCAGAAAGAAACTTATGGTCTAAAGAACCCACTTACTCTGATGAGCTATGTTACTCCAGTTATG |
| Microexon-tag Amino Acid seq | AVMSGFRWSMTQILLQKETYGLKNPLTLMSYVTPVM |
| Transcript ID | Ps.866.2 |
| Gene ID | Ps.866 |
| Gene Name | NA |
| Pfam domain motif | TPT |
| Motif E-value | 1.2e-29 |
| Motif start | 160 |
| Motif end | 459 |
| Protein seq | >Ps.866.2 MVDSNLEVEKDDAIELQDIQKESDRPVLRREPSFSRWFDEDGVIHFDRPLEHADETVEDSDFELPFLQTEGQESKLLSKD RLSNPRNMHLNGNHTMPMVRTHNHVDENTNGLNYVPFDIENESIRELPVSDFDAHDSSSRGSHKTTLDTQNSLDMAIVLK TLFFILVWYTFSTCLTIYNKTLLGEKLGKFPAPMLMNTIHFGMQAVLSNFITWFWSNRFQPTVTMSWKDYFMRVVPTALG TALDINLSNASLVFISITFATMCKSASPIFLLLFAFAFKLESPSVKLLGIISVISVGILLTVAKETEFQFWGFVFVMLAA VMSGFRWSMTQILLQKETYGLKNPLTLMSYVTPVMAVATAILSLAMDPWDEFRKNIYFDSSWHVFRSALLMFLGGTLAFF MVLTEYILVSATSAVTVTIAGVVKEVVTILVAVFYFHDKFTWLKGVGLFIIIFGVSLFNWYKYQKHQERQTSVVDESGLS PKNVAAKYVILEEMEDHDVDT* |
| CDS seq | >Ps.866.2 ATGGTAGACAGTAATTTGGAGGTGGAAAAGGATGATGCAATTGAGCTTCAAGATATTCAGAAAGAAAGTGACAGACCGGT TCTTCGAAGGGAACCATCTTTCTCACGCTGGTTTGACGAGGACGGCGTCATTCATTTCGATCGTCCTTTGGAGCATGCTG ATGAAACTGTAGAAGACTCTGATTTTGAGTTGCCGTTTCTTCAAACTGAAGGACAAGAGAGCAAATTATTGAGCAAGGAC CGACTGTCTAATCCGAGAAACATGCACCTGAATGGGAATCACACCATGCCCATGGTTAGGACTCATAATCATGTGGATGA AAATACAAATGGACTGAACTATGTGCCATTTGACATTGAAAATGAGTCTATTCGAGAATTGCCTGTTTCAGATTTTGATG CCCATGACTCCAGTTCAAGAGGAAGCCATAAAACGACACTGGATACACAGAATTCTCTTGATATGGCCATTGTGCTCAAG ACATTGTTTTTCATACTCGTATGGTACACTTTCAGTACATGTTTGACAATTTATAACAAAACATTATTAGGAGAAAAGTT GGGTAAATTCCCGGCTCCCATGTTGATGAATACTATCCACTTTGGGATGCAAGCTGTTCTATCGAATTTTATTACATGGT TTTGGTCTAACAGATTTCAACCCACAGTAACAATGTCATGGAAGGATTACTTCATGAGAGTTGTTCCCACGGCTCTTGGA ACTGCACTGGACATAAACCTGAGCAATGCCTCTCTTGTGTTCATCTCTATCACATTTGCCACCATGTGTAAATCAGCATC TCCTATATTTCTACTTCTATTTGCTTTTGCATTCAAGTTGGAGTCTCCAAGTGTTAAACTTCTTGGCATCATATCAGTCA TCTCTGTTGGAATTTTGTTAACAGTTGCAAAGGAAACAGAGTTTCAATTTTGGGGATTCGTATTTGTTATGCTTGCTGCT GTCATGTCTGGGTTTCGCTGGTCCATGACTCAGATTCTTTTGCAGAAAGAAACTTATGGTCTAAAGAACCCACTTACTCT GATGAGCTATGTTACTCCAGTTATGGCTGTGGCAACTGCTATACTCTCTCTTGCTATGGATCCATGGGATGAATTCAGAA AGAATATTTACTTTGATAGTTCATGGCATGTCTTTCGAAGTGCATTGTTGATGTTTCTTGGCGGAACGCTAGCCTTCTTT ATGGTTTTAACAGAATACATTCTTGTCTCAGCAACCAGTGCGGTAACTGTAACAATTGCAGGAGTCGTCAAGGAAGTTGT CACTATTTTGGTTGCGGTGTTTTACTTCCATGACAAATTTACCTGGTTGAAGGGAGTTGGGCTCTTCATAATAATCTTTG GTGTCAGTTTATTCAACTGGTACAAATATCAAAAGCATCAAGAACGCCAAACTAGTGTAGTTGATGAATCTGGGTTATCA CCCAAAAATGTAGCTGCGAAGTATGTTATCCTTGAGGAGATGGAGGATCACGATGTCGATACTTGA |