
| Microexon ID | Vv_10:1114709-1114720:+ |
| Species | Vistis vinifera | Coordinates | 10:1114709..1114720 |
| Microexon Cluster ID | MEP30 |
| Size | 12 |
| Phase | 1 |
| Pfam Domain Motif | SPARK |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,12,47 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | RATGTYTAYRRYCTTTGTCATATAASCCTSAARGAYTTYTCYCTYCAAGTTGGAWMWCAAGARTCTGGWTGYCTWYTDCCAAGYTTGCCTTCAGATGCAAYATTTGAY |
| Logo of Microexon-tag DNA Seq | 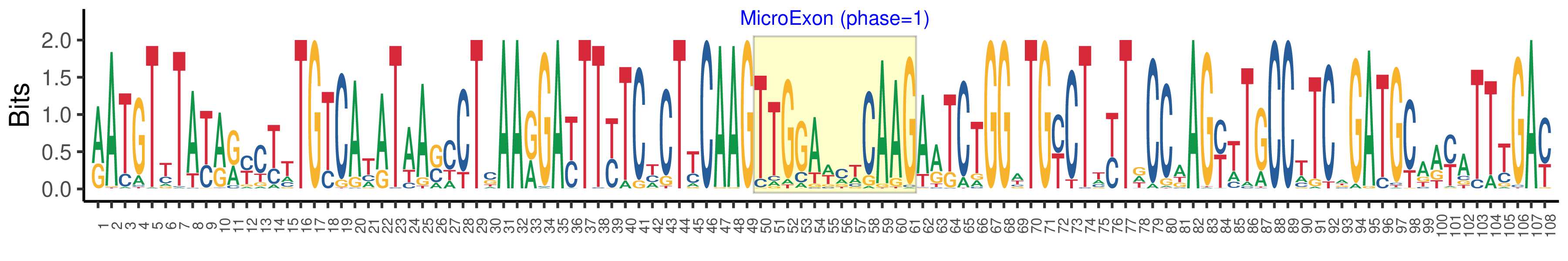 |
| Alignment of exons | 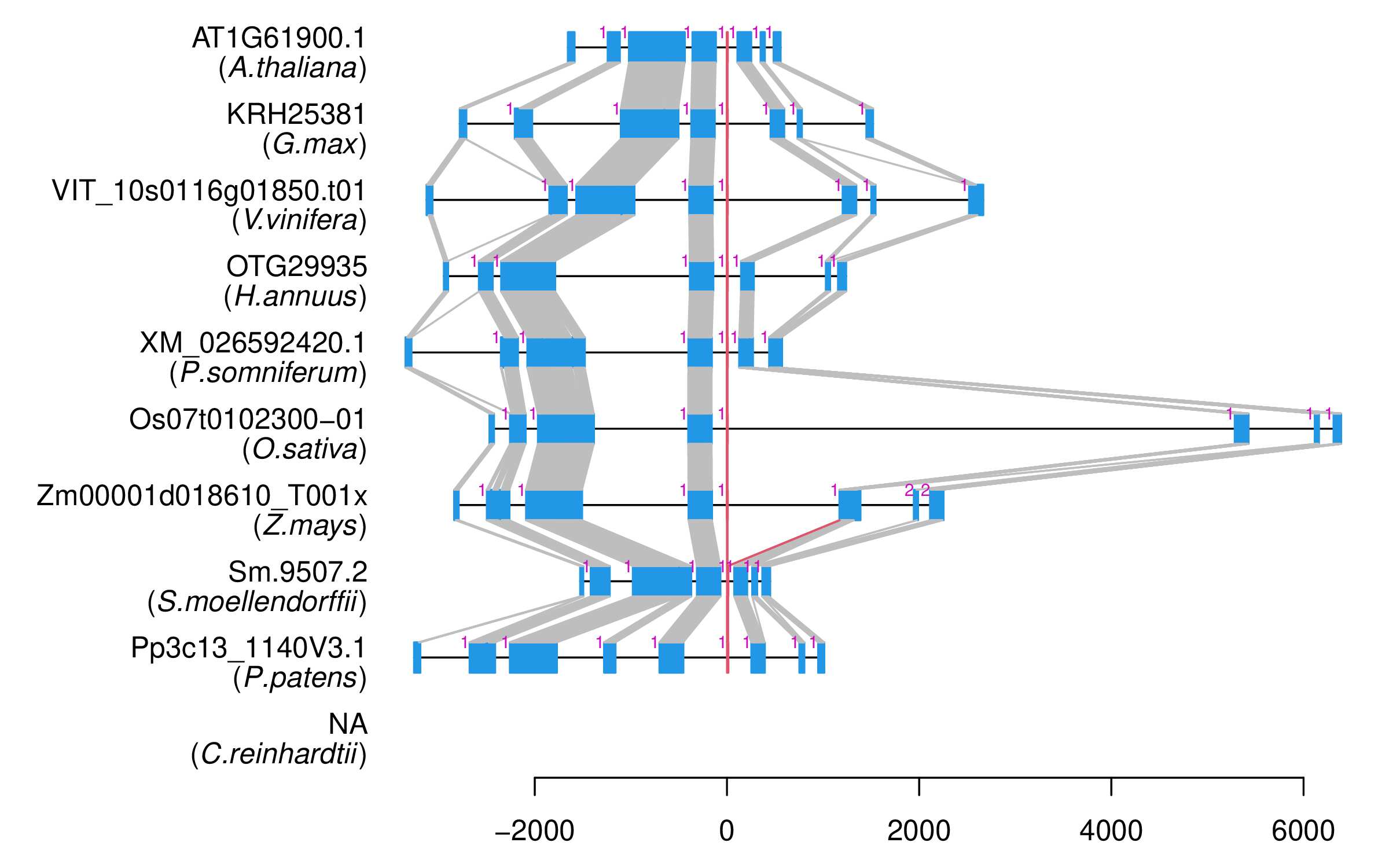 |
Vv_10:1114709-1114720:+ does not have available information here.
| Transcript ID | VIT_10s0116g01850.t01 |
| Protein ID | VIT_10s0116g01850.t01 |
| Gene ID | VIT_10s0116g01850 |
| Gene Name | NA |
| Pfam domain motif | Unknown |
| Motif E-value | NA |
| Motif start | NA |
| Motif end | NA |
| Protein seq | >VIT_10s0116g01850.t01 MRDGVCSNFSVLSVFLLVLFLCLHESHCSLPSYMKGSVLMEKSSDAFLPEISPSGAPQPFLPLLAPSPLTPFTTNSTIPK LSGLCSLNFSAGESMISMTSIDCWAVFASVLANVICCPQLKATLHILIGQSSKDTNLLALNETLAKHCLSDVEQILVSQG ASDKLHQICSIHPSSLTEGSCPVKDVNEFESIVDSTKLLSACEQIDLVKECCNQICQNAILEAARKIALKAYELLSVDPP HALPEHSTRVDDCKNIVLRWLASKLSPGRAKGVLRGLSNCNVNKVCPLVFPDTRHVAKGCGNGTSNQTACCSSMERYVSH LQKQSLITNLQALDCAASLGMKLQKANITRNIYNLCRITLKDFSLQAVGQESGCLLPSLPSDATLDKSSGISFICDLNDN IPAPWPSSSQLPASSCNKTVKIPALPAVASAQSDAPESVIQEGLKDGIKGYIISSALCTCVGSGRDAGGRVLDSKKCNQW NP* |
| CDS seq | >VIT_10s0116g01850.t01 ATGAGAGACGGGGTGTGTAGCAATTTCAGTGTGCTCAGTGTGTTTCTGTTGGTTCTGTTTCTCTGTCTTCATGAATCCCA TTGCAGCCTGCCAAGTTATATGAAAGGTTCTGTATTAATGGAAAAAAGCTCAGATGCCTTTTTACCTGAGATCTCCCCAA GTGGTGCTCCACAGCCATTTCTTCCTCTTCTTGCTCCTTCACCGCTGACACCATTCACAACAAATAGCACTATCCCAAAA TTATCAGGACTTTGTAGTTTAAACTTCTCTGCTGGTGAAAGTATGATCAGCATGACATCAATTGATTGCTGGGCTGTATT TGCTTCAGTTCTGGCTAATGTAATATGCTGTCCGCAGTTGAAAGCTACACTACACATTCTCATTGGCCAATCCAGCAAAG ACACCAATCTTCTCGCTTTGAATGAGACACTGGCAAAGCACTGCCTCTCGGATGTTGAGCAAATTCTAGTGAGTCAGGGT GCCAGTGATAAACTTCATCAGATATGCTCAATTCATCCATCAAGCCTCACTGAAGGCTCTTGCCCAGTCAAAGATGTTAA TGAGTTCGAGAGCATTGTGGATTCTACTAAGCTGCTTTCTGCATGTGAACAAATTGATCTTGTGAAAGAATGCTGTAATC AAATATGTCAGAATGCCATATTAGAAGCAGCTAGAAAAATCGCACTGAAAGCTTATGAGCTTTTGAGTGTGGATCCACCC CATGCACTGCCTGAACACTCGACTAGAGTTGATGACTGTAAAAATATTGTACTGCGATGGTTGGCAAGTAAGCTCAGTCC TGGCCGAGCCAAGGGTGTTCTTAGAGGACTATCTAATTGTAATGTTAATAAAGTTTGTCCTCTGGTTTTTCCTGACACAA GACATGTTGCAAAGGGCTGTGGAAATGGGACAAGCAACCAGACGGCATGCTGTAGTTCTATGGAGAGGTATGTGTCTCAT TTGCAAAAGCAGAGTTTGATAACCAACTTGCAAGCTTTGGACTGTGCTGCATCACTTGGAATGAAGTTACAGAAGGCAAA TATTACGAGAAATATTTATAACCTCTGTCGAATTACCCTCAAGGATTTCTCTCTCCAAGCCGTTGGACAAGAGTCTGGAT GCCTTTTGCCAAGCTTGCCTTCAGATGCAACATTGGACAAGTCTTCAGGGATAAGTTTCATTTGTGATCTGAATGACAAC ATTCCAGCTCCTTGGCCTTCTTCATCTCAATTACCAGCTTCATCATGCAATAAAACTGTCAAAATTCCTGCACTTCCTGC AGTCGCATCTGCTCAAAGTGATGCTCCTGAATCAGTTATCCAGGAGGGGCTAAAGGATGGTATTAAGGGATACATCATAA GCAGTGCCTTGTGCACCTGTGTTGGCAGTGGAAGAGATGCCGGTGGGAGAGTTCTTGATAGCAAAAAATGCAATCAGTGG AACCCTTGA |
| Microexon DNA seq | CCGTTGGACAAG |
| Microexon Amino Acid seq | AVGQE |
| Microexon-tag DNA Seq | AATATTTATAACCTCTGTCGAATTACCCTCAAGGATTTCTCTCTCCAAGCCGTTGGACAAGAGTCTGGATGCCTTTTGCCAAGCTTGCCTTCAGATGCAACATTGGAC |
| Microexon-tag Amino Acid seq | NIYNLCRITLKDFSLQAVGQESGCLLPSLPSDATLD |
| Transcript ID | Vv.1951.1 |
| Gene ID | Vv.1951 |
| Gene Name | NA |
| Pfam domain motif | Unknown |
| Motif E-value | NA |
| Motif start | NA |
| Motif end | NA |
| Protein seq | >Vv.1951.1 MRDGVCSNFSVLSVFLLVLFLCLHESHCSLPSYMKGSVLMEKSSDAFLPEISPSGAPQPFLPLLAPSPLTPFTTNSTIPK LSGLCSLNFSAGESMISMTSIDCWAVFASVLANVICCPQLKATLHILIGQSSKDTNLLALNETLAKHCLSDVEQILVSQG ASDKLHQICSIHPSSLTEGSCPVKDVNEFESIVDSTKLLSACEQIDLVKECCNQICQNAILEAARKIALKAYELLSVDPP HALPEHSTRVDDCKNIVLRWLASKLSPGRAKGVLRGLSNCNVNKVCPLVFPDTRHVAKGCGNGTSNQTACCSSMERYVSH LQKQSLITNLQALDCAASLGMKLQKANITRNIYNLCRITLKDFSLQAVGQESGCLLPSLPSDATLDKSSGISFICDLNDN IPAPWPSSSQLPASSCNKTVKIPALPAVASAQSGLYSEDIMPRLLFTSSMVLTMLL* |
| CDS seq | >Vv.1951.1 ATGAGAGACGGGGTGTGTAGCAATTTCAGTGTGCTCAGTGTGTTTCTGTTGGTTCTGTTTCTCTGTCTTCATGAATCCCA TTGCAGCCTGCCAAGTTATATGAAAGGTTCTGTATTAATGGAAAAAAGCTCAGATGCCTTTTTACCTGAGATCTCCCCAA GTGGTGCTCCACAGCCATTTCTTCCTCTTCTTGCTCCTTCACCGCTGACACCATTCACAACAAATAGCACTATCCCAAAA TTATCAGGACTTTGTAGTTTAAACTTCTCTGCTGGTGAAAGTATGATCAGCATGACATCAATTGATTGCTGGGCTGTATT TGCTTCAGTTCTGGCTAATGTAATATGCTGTCCGCAGTTGAAAGCTACACTACACATTCTCATTGGCCAATCCAGCAAAG ACACCAATCTTCTCGCTTTGAATGAGACACTGGCAAAGCACTGCCTCTCGGATGTTGAGCAAATTCTAGTGAGTCAGGGT GCCAGTGATAAACTTCATCAGATATGCTCAATTCATCCATCAAGCCTCACTGAAGGCTCTTGCCCAGTCAAAGATGTTAA TGAGTTCGAGAGCATTGTGGATTCTACTAAGCTGCTTTCTGCATGTGAACAAATTGATCTTGTGAAAGAATGCTGTAATC AAATATGTCAGAATGCCATATTAGAAGCAGCTAGAAAAATCGCACTGAAAGCTTATGAGCTTTTGAGTGTGGATCCACCC CATGCACTGCCTGAACACTCGACTAGAGTTGATGACTGTAAAAATATTGTACTGCGATGGTTGGCAAGTAAGCTCAGTCC TGGCCGAGCCAAGGGTGTTCTTAGAGGACTATCTAATTGTAATGTTAATAAAGTTTGTCCTCTGGTTTTTCCTGACACAA GACATGTTGCAAAGGGCTGTGGAAATGGGACAAGCAACCAGACGGCATGCTGTAGTTCTATGGAGAGGTATGTGTCTCAT TTGCAAAAGCAGAGTTTGATAACCAACTTGCAAGCTTTGGACTGTGCTGCATCACTTGGAATGAAGTTACAGAAGGCAAA TATTACGAGAAATATTTATAACCTCTGTCGAATTACCCTCAAGGATTTCTCTCTCCAAGCCGTTGGACAAGAGTCTGGAT GCCTTTTGCCAAGCTTGCCTTCAGATGCAACATTGGACAAGTCTTCAGGGATAAGTTTCATTTGTGATCTGAATGACAAC ATTCCAGCTCCTTGGCCTTCTTCATCTCAATTACCAGCTTCATCATGCAATAAAACTGTCAAAATTCCTGCACTTCCTGC AGTCGCATCTGCTCAAAGTGGTCTTTACAGTGAAGATATAATGCCACGACTGCTCTTTACCTCCTCTATGGTTCTCACGA TGCTCCTGTAA |