
| Microexon ID | Sl_3:4172635-4172646:+ |
| Species | Solanum Lycopersicum | Coordinates | 3:4172635..4172646 |
| Microexon Cluster ID | MEP30 |
| Size | 12 |
| Phase | 1 |
| Pfam Domain Motif | SPARK |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,12,47 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | RATGTYTAYRRYCTTTGTCATATAASCCTSAARGAYTTYTCYCTYCAAGTTGGAWMWCAAGARTCTGGWTGYCTWYTDCCAAGYTTGCCTTCAGATGCAAYATTTGAY |
| Logo of Microexon-tag DNA Seq | 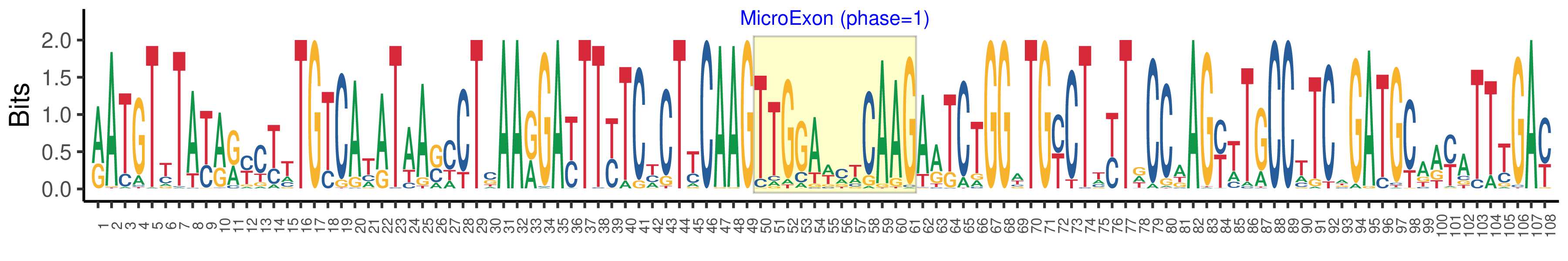 |
| Alignment of exons | 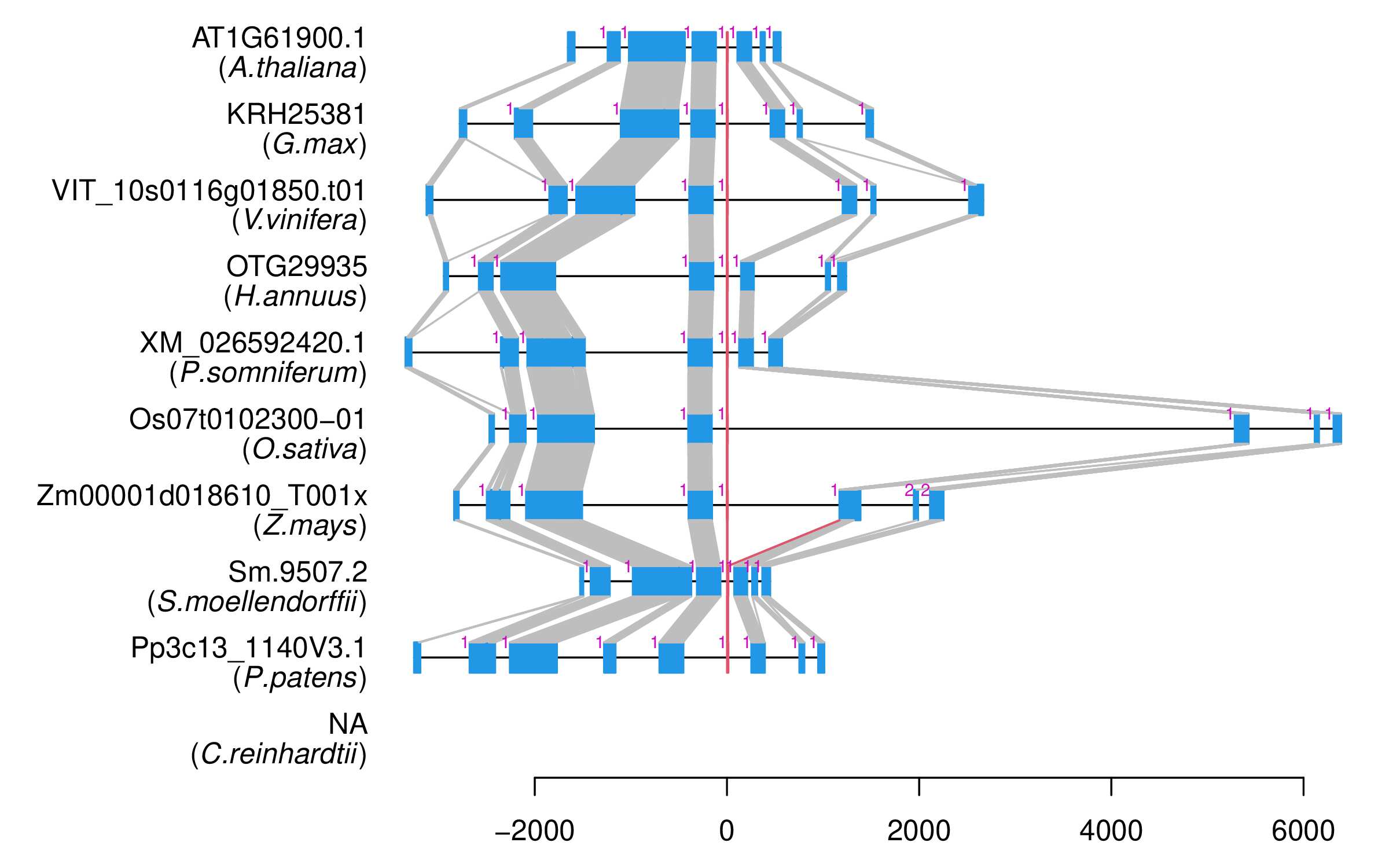 |
| Microexon DNA seq | TTACATCAGAAG |
| Microexon Amino Acid seq | VTSEV |
| Microexon-tag DNA Seq | AATGTCTACAATCTCTGTCACATTAGCCTCAAGGATTTCTCCGTACAAGTTACATCAGAAGTTTCGGGGTGTCTTTTGTCTAGTTTACCATCGGATGCAATACTGGAC |
| Microexon-tag Amino Acid Seq | NVYNLCHISLKDFSVQVTSEVSGCLLSSLPSDAILD |
| Microexon-tag spanning region | 4172462-4173525 |
| Microexon-tag prediction score | 0.9288 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | Solyc03g031680.3.1x |
| Reference Transcript ID | Solyc03g031680.3.1 |
| Gene ID | Solyc03g031680.3 |
| Gene Name | NA |
| Transcript ID | Solyc03g031680.3.1 |
| Protein ID | |
| Gene ID | Solyc03g031680.3 |
| Gene Name | |
| Pfam domain motif | Unknown |
| Motif E-value | NA |
| Motif start | NA |
| Motif end | NA |
| Protein seq | >Solyc03g031680.3.1 MRGGVSESFRLHLLLVVFILFLCLQESSCSSLNHLRSSLIMDIKEDTLLLETLPTAAPQPLLPLRAPSPLAPFTNSTSLR LSGLCTLNFDAVKSMMAVTSIDCVAPFAEYLANVICCPQLETTLVILIGQSSKHSNMLALNGTLAKHCLSDFQQLLVSQG ANDTLQKICSLHPENLTEGSCPVKDVHEFESTVDSSSLLAACGKIDLVNECCEETCGNAITEAAKKLALKAYDISRMGGP HVLSDHTTRVNDCKRIVQRWLANKLEPAGAKHVLRGLYNCKNNKVCPLVFPNMKNITKACGDGMNNQTRCCNTVERYVSH LQRQSFVTNLQALDCAASLGLKLQKDNVSKNVYNLCHISLKDFSVQVTSEVSGCLLSSLPSDAILDKTVGISFVCDLNDN IPAPWPSMSQLPASSCKKSVRIPALPAVASGQISFRGLNIWSYLLLAASMILEVCCIYNAAILPY* |
| CDS seq | >Solyc03g031680.3.1 ATGAGAGGAGGGGTCAGTGAAAGTTTCAGACTTCACCTTCTTCTGGTTGTATTTATACTCTTTCTTTGCCTCCAGGAATC CAGTTGCAGCTCACTAAATCATTTAAGAAGCTCTCTAATCATGGACATAAAGGAGGATACCCTTTTGCTAGAGACGTTAC CAACTGCTGCTCCACAACCCCTTCTCCCCCTTCGTGCACCTTCTCCATTGGCACCTTTCACAAACAGCACTTCACTTAGA CTATCTGGACTGTGTACGCTGAACTTTGATGCTGTGAAAAGTATGATGGCCGTGACATCAATAGATTGTGTAGCTCCATT TGCAGAGTATCTGGCTAATGTCATCTGTTGCCCTCAACTAGAAACAACTCTTGTTATTCTTATTGGGCAGTCTAGTAAAC ATTCAAATATGCTTGCTTTAAATGGGACTCTCGCAAAGCACTGCCTTTCAGATTTTCAGCAACTCCTGGTCAGTCAGGGT GCCAATGATACTCTGCAGAAGATATGCTCTCTCCATCCGGAAAACCTTACTGAAGGTTCTTGCCCAGTCAAAGATGTTCA TGAGTTTGAGAGCACTGTGGACTCCTCTAGCCTACTTGCTGCCTGTGGCAAGATCGATCTTGTGAATGAATGTTGTGAGG AAACTTGTGGAAATGCTATAACAGAAGCTGCTAAAAAACTTGCACTTAAAGCATATGATATTTCAAGGATGGGTGGCCCT CATGTGCTGTCTGATCACACAACTAGAGTTAACGACTGTAAAAGGATTGTACAAAGATGGCTGGCAAATAAACTTGAACC TGCTGGAGCAAAACATGTTCTTAGAGGACTTTATAATTGCAAAAACAATAAAGTGTGCCCTTTGGTTTTTCCTAACATGA AAAATATTACAAAGGCTTGTGGAGATGGGATGAATAATCAAACAAGATGCTGTAATACTGTCGAGAGGTATGTGTCTCAC TTACAGAGGCAGAGCTTTGTCACCAACTTGCAAGCTTTGGATTGTGCTGCCTCGCTTGGGCTTAAGCTACAGAAAGACAA TGTTAGCAAAAATGTCTACAATCTCTGTCACATTAGCCTCAAGGATTTCTCCGTACAAGTTACATCAGAAGTTTCGGGGT GTCTTTTGTCTAGTTTACCATCGGATGCAATACTGGACAAAACTGTGGGGATCAGCTTTGTATGCGACTTAAATGACAAC ATTCCGGCTCCTTGGCCATCTATGTCTCAGTTACCAGCTTCATCATGCAAGAAGTCTGTGAGGATTCCTGCACTTCCTGC TGTAGCATCGGGGCAAATAAGTTTTAGAGGATTAAATATATGGTCATACCTTCTGCTGGCGGCCTCGATGATACTAGAAG TCTGCTGTATATATAATGCTGCCATTCTTCCTTACTAG |
Sl_3:4172635-4172646:+ does not have available information here.