
| Microexon ID | Sb_2:307885-307896:- |
| Species | Sorghum Bicolor | Coordinates | 2:307885..307896 |
| Microexon Cluster ID | MEP30 |
| Size | 12 |
| Phase | 1 |
| Pfam Domain Motif | SPARK |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,12,47 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | RATGTYTAYRRYCTTTGTCATATAASCCTSAARGAYTTYTCYCTYCAAGTTGGAWMWCAAGARTCTGGWTGYCTWYTDCCAAGYTTGCCTTCAGATGCAAYATTTGAY |
| Logo of Microexon-tag DNA Seq | 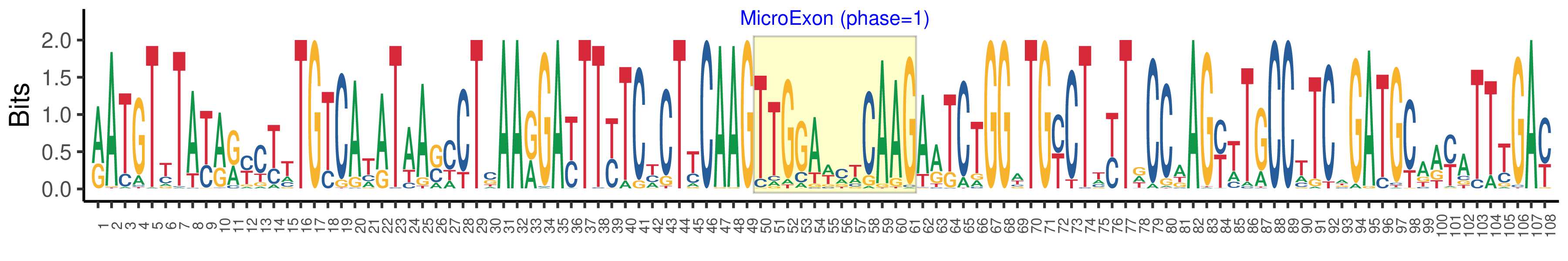 |
| Alignment of exons |  |
| Microexon DNA seq | TTGGATCTCAAG |
| Microexon Amino Acid seq | VGSQE |
| Microexon-tag DNA Seq | AATGTCTATAGCTCTTGTCAAATAACACTAAAAGATTTTTCACTTCAAGTTGGATCTCAAGAATCTGGATGCCTCCTTCCAAGTATGCCCTCTGATGCATCTTTCGAC |
| Microexon-tag Amino Acid Seq | NVYSSCQITLKDFSLQVGSQESGCLLPSMPSDASFD |
| Microexon-tag spanning region | 307237-308096 |
| Microexon-tag prediction score | 0.9684 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | EER97817x |
| Reference Transcript ID | EER97817 |
| Gene ID | SORBI_3002G002100 |
| Gene Name | NA |
| Transcript ID | EER97817 |
| Protein ID | |
| Gene ID | SORBI_3002G002100 |
| Gene Name | |
| Pfam domain motif | Unknown |
| Motif E-value | NA |
| Motif start | NA |
| Motif end | NA |
| Protein seq | >EER97817 MAPAGRLLAVAALSFLCLQGLWGDSGSALAQSQPPLHRIVRAEANDGGGGGLMPELPPSGFMPMPDLSPSPSGSPRPFVP FLAPAPLAPFFNNSTPKLSGKCTLNFTAVDKLMTTTAVDCFTSFAPFLANVICCPQLQATLTILIGQSSKQTGSLALDPT VANYCLSDVQELLLSQGASDNLHSLCSVHLSNVTEGSCPVSTVDSFESVVDSSKLLEACRKIDSVNECCSQTCQNAINEA AQKISSKDGGLTTYPGSPKVDSCRNVVLRWLSSRLDPQSAKEMLRQISNCNVNGVCPLSFPDTSKVAKECGGTMKNSTAC CKAMLTYVAHLQKQSFITNLQALNCASFLGAKLQKMNVSTNVYSSCQITLKDFSLQVGSQESGCLLPSMPSDASFDSTSG ISFTCDLNDNIAAPWPSSMQAPSSSCNKSVNIPERPAATSAQNGVNHKNLKLRLLASLVSLLLVLLVQV* |
| CDS seq | >EER97817 ATGGCGCCTGCCGGGCGGCTCCTCGCGGTAGCCGCCCTCTCCTTCCTCTGTTTGCAGGGGCTGTGGGGCGATTCAGGCAG CGCATTGGCTCAGAGCCAGCCGCCGCTTCACCGGATCGTTCGCGCAGAGGCAAATGACGGCGGCGGCGGCGGCCTCATGC CGGAGCTCCCGCCCAGTGGCTTCATGCCCATGCCGGACCTCTCACCCTCACCCAGCGGCTCCCCGAGGCCATTTGTTCCC TTCCTGGCTCCCGCACCACTCGCGCCCTTCTTTAACAACAGCACGCCAAAGCTCTCAGGGAAATGCACACTCAACTTTAC CGCTGTCGACAAACTCATGACCACCACGGCTGTCGACTGCTTCACCTCGTTTGCACCTTTCTTGGCCAATGTCATCTGCT GCCCGCAGCTTCAGGCAACACTGACCATCCTCATAGGGCAGTCTAGCAAGCAGACAGGGTCTCTCGCATTGGATCCCACG GTGGCCAACTACTGCCTGTCGGATGTCCAGGAGCTGTTGTTGAGCCAAGGCGCCAGTGATAATCTCCATAGCCTTTGCTC GGTTCACCTCTCGAATGTAACTGAGGGGTCCTGCCCTGTTAGCACTGTGGATTCGTTTGAGAGCGTCGTTGATTCGTCCA AGCTCCTTGAGGCCTGTCGGAAGATCGACTCTGTTAATGAGTGCTGCAGCCAGACATGCCAGAATGCGATAAATGAAGCT GCCCAGAAAATCTCGTCTAAGGATGGAGGTTTGACGACCTACCCTGGGAGCCCCAAGGTTGACAGTTGCAGGAACGTAGT TCTGAGATGGTTGTCAAGCAGGCTTGACCCACAGTCCGCGAAGGAAATGTTGAGGCAGATATCTAACTGTAATGTCAATG GAGTTTGTCCGCTAAGTTTTCCAGACACAAGTAAGGTGGCAAAAGAGTGTGGTGGAACGATGAAGAACAGTACTGCATGT TGCAAGGCTATGTTAACTTATGTAGCCCATTTGCAAAAGCAAAGCTTCATAACAAACTTGCAAGCTTTAAATTGTGCTTC TTTCCTTGGGGCAAAGTTGCAAAAGATGAATGTTAGCACGAATGTCTATAGCTCTTGTCAAATAACACTAAAAGATTTTT CACTTCAAGTTGGATCTCAAGAATCTGGATGCCTCCTTCCAAGTATGCCCTCTGATGCATCTTTCGACAGCACTTCTGGG ATTAGCTTCACATGTGATCTGAATGACAACATTGCTGCTCCATGGCCATCTTCCATGCAAGCACCATCTTCATCATGCAA CAAGTCTGTCAATATCCCAGAGCGACCCGCTGCGACATCGGCACAAAATGGTGTGAACCACAAGAACCTGAAGCTAAGGC TGCTTGCTTCTTTGGTCTCCCTTCTTCTGGTGCTCTTGGTACAAGTTTAG |
Sb_2:307885-307896:- does not have available information here.