
| Microexon ID | At_1:22883059-22883070:- |
| Species | Arabidopsis thaliana | Coordinates | 1:22883059..22883070 |
| Microexon Cluster ID | MEP30 |
| Size | 12 |
| Phase | 1 |
| Pfam Domain Motif | SPARK |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,12,47 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | RATGTYTAYRRYCTTTGTCATATAASCCTSAARGAYTTYTCYCTYCAAGTTGGAWMWCAAGARTCTGGWTGYCTWYTDCCAAGYTTGCCTTCAGATGCAAYATTTGAY |
| Logo of Microexon-tag DNA Seq | 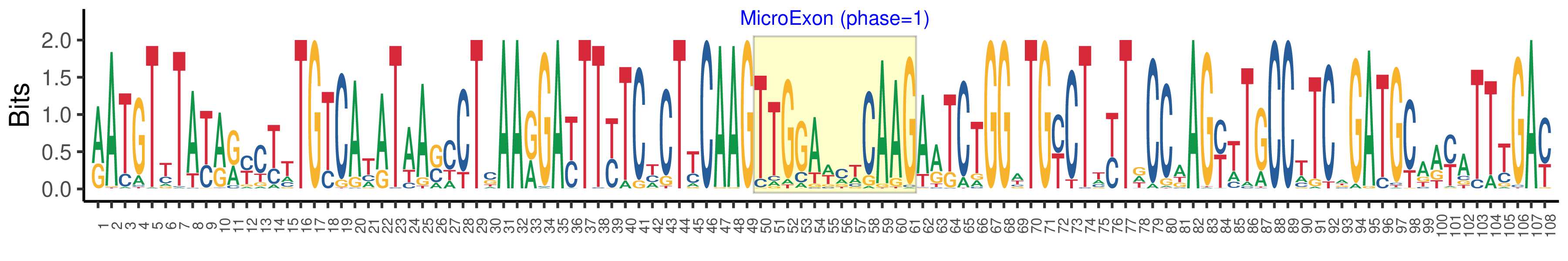 |
| Alignment of exons | 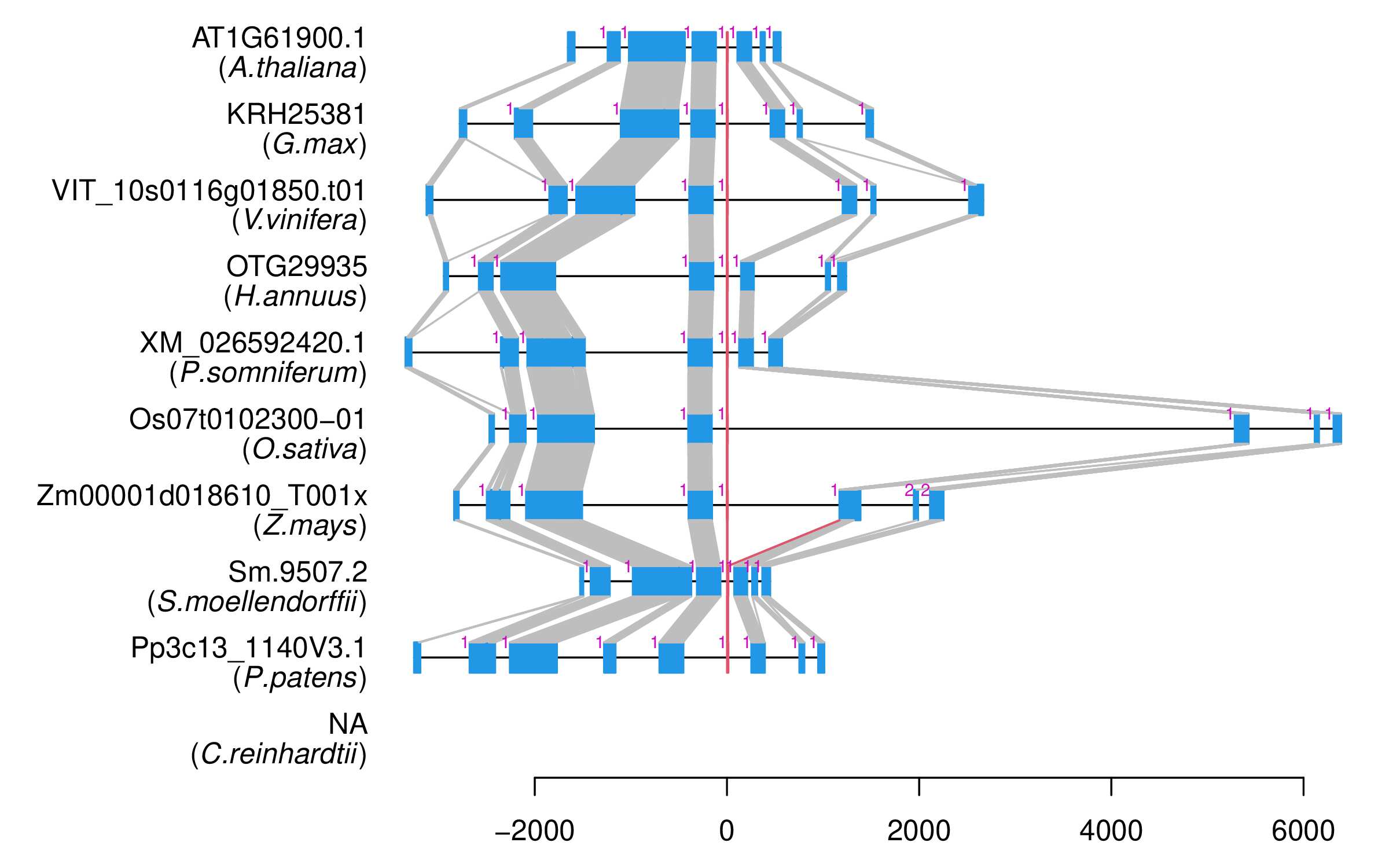 |
| Microexon DNA seq | TTGGAAACCAAG |
| Microexon Amino Acid seq | VGNQE |
| Microexon-tag DNA Seq | AACATATTCAGTGTTTGTCATATAAGTCTCAAGGACTTCTCTCTTCAAGTTGGAAACCAAGAATCTGGTTGTCTATTACCGAGCCTACCATCGGATGCGATATTTGAT |
| Microexon-tag Amino Acid Seq | NIFSVCHISLKDFSLQVGNQESGCLLPSLPSDAIFD |
| Microexon-tag spanning region | 22882912-22883230 |
| Microexon-tag prediction score | 0.9553 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | AT1G61900.1x |
| Reference Transcript ID | AT1G61900.1 |
| Gene ID | AT1G61900 |
| Gene Name | NA |
| Transcript ID | AT1G61900.1 |
| Protein ID | AT1G61900.1 |
| Gene ID | AT1G61900 |
| Gene Name | NA |
| Pfam domain motif | SPARK |
| Motif E-value | 0.24 |
| Motif start | 261 |
| Motif end | 350 |
| Protein seq | >AT1G61900.1 MTRRAEFEMGLFVILQSMFLISLCSSQKPEEFLPEISPDTSPQPFLPFIAPSPMVPYINSTMPKLSGLCSLNFSASESLI QTTSHNCWTVFAPLLANVMCCPQLDATLTIILGKASKETGLLALNRTQSKHCLSDLEQILVGKGASGQLNKICSIHSSNL TSSSCPVINVDEFESTVDTAKLLLACEKIDPVKECCEEACQNAILDAATNISLKASETLTDNSDRINDCKNVVNRWLATK LDPSRVKETLRGLANCKINRVCPLVFPHMKHIGGNCSNELSNQTGCCRAMESYVSHLQKQTLITNLQALDCATSLGTKLQ KLNITKNIFSVCHISLKDFSLQVGNQESGCLLPSLPSDAIFDKDTGISFTCDLNDNIPAPWPSSSLSSASTCKKPVRIPA LPAAASSQPRLHDEGVTRLVIFVLSMLLVMLLS* |
| CDS seq | >AT1G61900.1 ATGACGAGAAGAGCTGAATTTGAGATGGGTTTGTTCGTGATTCTTCAATCCATGTTTCTGATATCTCTATGTTCATCACA GAAACCTGAGGAGTTCTTACCTGAGATATCACCAGATACTTCTCCTCAACCGTTTCTTCCTTTCATTGCACCTTCTCCAA TGGTTCCATACATAAATAGTACCATGCCTAAACTATCAGGGCTTTGCTCGCTTAATTTCAGTGCGTCTGAGAGTTTGATT CAGACTACTTCACACAACTGTTGGACTGTTTTTGCTCCATTGTTGGCAAATGTGATGTGTTGTCCTCAGCTAGATGCTAC ACTCACAATAATCCTTGGTAAAGCGAGTAAAGAAACCGGTTTGCTTGCTTTAAACAGAACCCAGTCTAAGCACTGTCTCT CGGATTTAGAGCAAATCTTGGTAGGAAAAGGCGCTTCAGGTCAGCTTAATAAGATCTGTTCTATTCACTCGTCGAATCTC ACTTCTTCGTCGTGCCCTGTTATCAATGTCGATGAGTTTGAAAGCACGGTGGATACAGCGAAACTTCTTTTGGCCTGTGA AAAAATTGATCCTGTGAAGGAATGTTGCGAAGAGGCTTGCCAAAATGCGATACTCGATGCTGCTACGAATATATCTCTTA AAGCATCAGAAACTTTAACTGATAATTCGGATAGGATCAATGACTGCAAGAACGTAGTAAACCGCTGGCTCGCTACTAAA CTTGACCCTTCTCGGGTTAAGGAAACTCTCAGAGGATTAGCAAACTGCAAAATCAATAGAGTTTGTCCTCTTGTCTTTCC GCACATGAAGCATATTGGTGGTAACTGCAGCAACGAGTTGAGTAACCAAACGGGTTGTTGCCGTGCAATGGAGAGTTATG TGTCTCACTTGCAAAAGCAAACACTTATAACCAATTTGCAAGCTTTAGATTGCGCAACCAGCCTTGGGACAAAGTTACAG AAACTGAACATCACTAAGAACATATTCAGTGTTTGTCATATAAGTCTCAAGGACTTCTCTCTTCAAGTTGGAAACCAAGA ATCTGGTTGTCTATTACCGAGCCTACCATCGGATGCGATATTTGATAAAGACACGGGGATAAGCTTTACTTGTGACCTCA ATGACAACATTCCCGCTCCGTGGCCTTCATCTTCTCTATCCTCAGCCTCTACCTGTAAAAAACCTGTTAGAATTCCTGCT CTTCCAGCTGCTGCCTCCTCGCAACCTCGTCTCCACGATGAAGGAGTGACACGTTTGGTGATCTTTGTCTTGTCTATGCT TCTTGTGATGCTTCTGTCATAG |
| Microexon DNA seq | TTGGAAACCAAG |
| Microexon Amino Acid seq | VGNQE |
| Microexon-tag DNA Seq | AACATATTCAGTGTTTGTCATATAAGTCTCAAGGACTTCTCTCTTCAAGTTGGAAACCAAGAATCTGGTTGTCTATTACCGAGCCTACCATCGGATGCGATATTTGAT |
| Microexon-tag Amino Acid seq | NIFSVCHISLKDFSLQVGNQESGCLLPSLPSDAIFD |
| Transcript ID | AT1G61900.1 |
| Gene ID | At.5295 |
| Gene Name | NA |
| Pfam domain motif | SPARK |
| Motif E-value | 0.24 |
| Motif start | 261 |
| Motif end | 350 |
| Protein seq | >AT1G61900.1 MTRRAEFEMGLFVILQSMFLISLCSSQKPEEFLPEISPDTSPQPFLPFIAPSPMVPYINSTMPKLSGLCSLNFSASESLI QTTSHNCWTVFAPLLANVMCCPQLDATLTIILGKASKETGLLALNRTQSKHCLSDLEQILVGKGASGQLNKICSIHSSNL TSSSCPVINVDEFESTVDTAKLLLACEKIDPVKECCEEACQNAILDAATNISLKASETLTDNSDRINDCKNVVNRWLATK LDPSRVKETLRGLANCKINRVCPLVFPHMKHIGGNCSNELSNQTGCCRAMESYVSHLQKQTLITNLQALDCATSLGTKLQ KLNITKNIFSVCHISLKDFSLQVGNQESGCLLPSLPSDAIFDKDTGISFTCDLNDNIPAPWPSSSLSSASTCKKPVRIPA LPAAASSQPRLHDEGVTRLVIFVLSMLLVMLLS* |
| CDS seq | >AT1G61900.1 ATGACGAGAAGAGCTGAATTTGAGATGGGTTTGTTCGTGATTCTTCAATCCATGTTTCTGATATCTCTATGTTCATCACA GAAACCTGAGGAGTTCTTACCTGAGATATCACCAGATACTTCTCCTCAACCGTTTCTTCCTTTCATTGCACCTTCTCCAA TGGTTCCATACATAAATAGTACCATGCCTAAACTATCAGGGCTTTGCTCGCTTAATTTCAGTGCGTCTGAGAGTTTGATT CAGACTACTTCACACAACTGTTGGACTGTTTTTGCTCCATTGTTGGCAAATGTGATGTGTTGTCCTCAGCTAGATGCTAC ACTCACAATAATCCTTGGTAAAGCGAGTAAAGAAACCGGTTTGCTTGCTTTAAACAGAACCCAGTCTAAGCACTGTCTCT CGGATTTAGAGCAAATCTTGGTAGGAAAAGGCGCTTCAGGTCAGCTTAATAAGATCTGTTCTATTCACTCGTCGAATCTC ACTTCTTCGTCGTGCCCTGTTATCAATGTCGATGAGTTTGAAAGCACGGTGGATACAGCGAAACTTCTTTTGGCCTGTGA AAAAATTGATCCTGTGAAGGAATGTTGCGAAGAGGCTTGCCAAAATGCGATACTCGATGCTGCTACGAATATATCTCTTA AAGCATCAGAAACTTTAACTGATAATTCGGATAGGATCAATGACTGCAAGAACGTAGTAAACCGCTGGCTCGCTACTAAA CTTGACCCTTCTCGGGTTAAGGAAACTCTCAGAGGATTAGCAAACTGCAAAATCAATAGAGTTTGTCCTCTTGTCTTTCC GCACATGAAGCATATTGGTGGTAACTGCAGCAACGAGTTGAGTAACCAAACGGGTTGTTGCCGTGCAATGGAGAGTTATG TGTCTCACTTGCAAAAGCAAACACTTATAACCAATTTGCAAGCTTTAGATTGCGCAACCAGCCTTGGGACAAAGTTACAG AAACTGAACATCACTAAGAACATATTCAGTGTTTGTCATATAAGTCTCAAGGACTTCTCTCTTCAAGTTGGAAACCAAGA ATCTGGTTGTCTATTACCGAGCCTACCATCGGATGCGATATTTGATAAAGACACGGGGATAAGCTTTACTTGTGACCTCA ATGACAACATTCCCGCTCCGTGGCCTTCATCTTCTCTATCCTCAGCCTCTACCTGTAAAAAACCTGTTAGAATTCCTGCT CTTCCAGCTGCTGCCTCCTCGCAACCTCGTCTCCACGATGAAGGAGTGACACGTTTGGTGATCTTTGTCTTGTCTATGCT TCTTGTGATGCTTCTGTCATAG |