
| Microexon ID | Ps_NC_039362.1:122173130-122173141:- |
| Species | Papaver somniferum | Coordinates | NC_039362.1:122173130..122173141 |
| Microexon Cluster ID | MEP29 |
| Size | 12 |
| Phase | 0 |
| Pfam Domain Motif | TPR_12 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,12,48 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TTYTATCTTRTSCARMRSAARYTKGAWSAAGCYCRKRCWTGYTATGAGCGTGCTYTRAAGATAAARRGRCGTGTTYTGGGRSWTRRYCAYSCAGAYTATGCARAKACA |
| Logo of Microexon-tag DNA Seq | 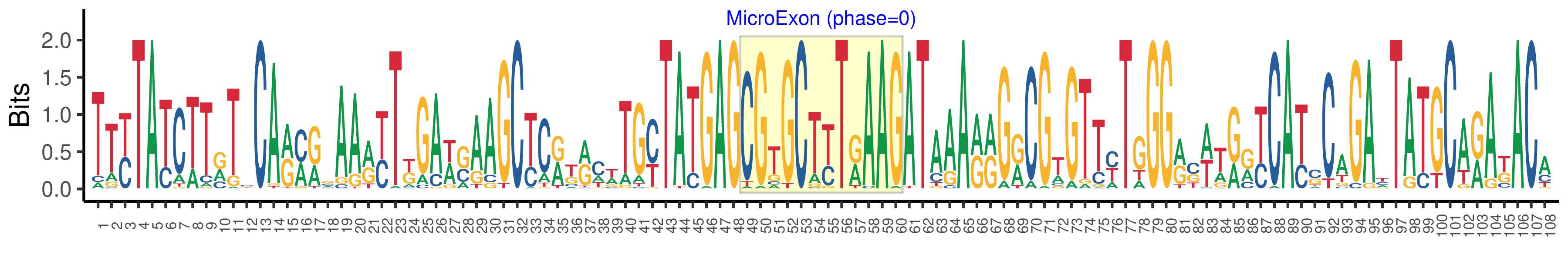 |
| Alignment of exons | 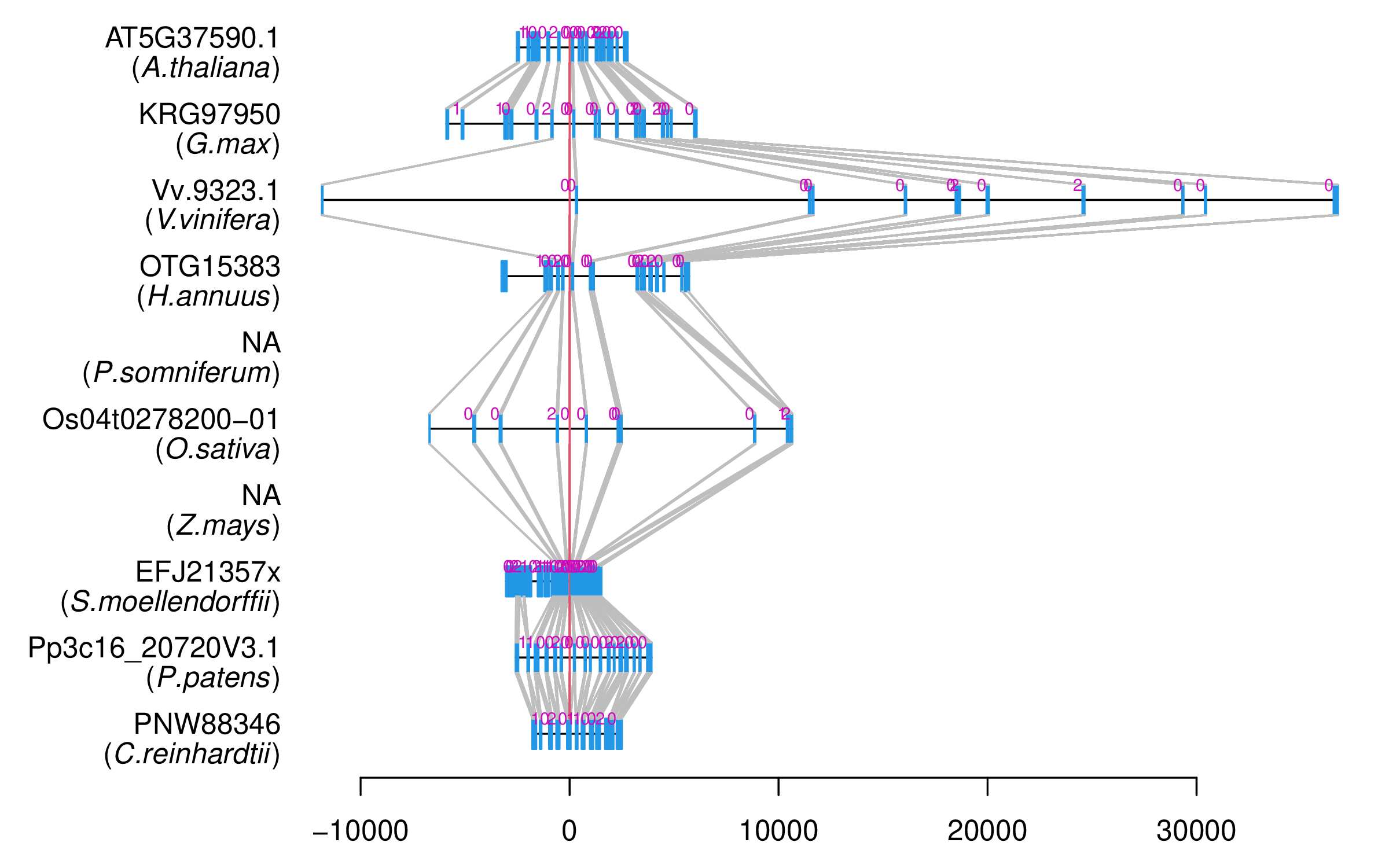 |
| Microexon DNA seq | CGTGCGTTAAAG |
| Microexon Amino Acid seq | RALK |
| Microexon-tag DNA Seq | TTTTATATTGTGCAGAGGAAACTTGAAGAAGCCCATAACTGCTACGAGCGTGCGTTAAAGATAAAGAGGCGTGTTCTTGGACAAAACCACACAGACTATGCAAATACA |
| Microexon-tag Amino Acid Seq | FYIVQRKLEEAHNCYERALKIKRRVLGQNHTDYANT |
| Microexon-tag spanning region | 122172867-122173817 |
| Microexon-tag prediction score | 0.9553 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | XM_026531570.1x |
| Reference Transcript ID | XM_026531570.1 |
| Gene ID | NA |
| Gene Name | NA |
| Transcript ID | XM_026531570.1 |
| Protein ID | XP_026387355.1 |
| Gene ID | LOC113282544 |
| Gene Name | NA |
| Pfam domain motif | TPR_12 |
| Motif E-value | 3.2e-14 |
| Motif start | 205 |
| Motif end | 280 |
| Protein seq | >XP_026387355.1 MFLRQSGSLLRRRLRYLLSTNTITTTTKTTISSLSSYPPPSNAGFLHHNGPEIPRWQKLHWDSSLWILLSGQAAIILGAS GNLAFADDVSAEARTGAGEDGSSDLQRVEDGSVISNIHTSKWRVFTDKARDFFLEGKLVEAEKLFLSAVEEAKEGFGKSD PHVAAALNNLAELYRVKKDFAKAEPLYLEAVHILEESFGLGDIRVAASLHNLGQFYIVQRKLEEAHNCYERALKIKRRVL GQNHTDYANTMYHLGTVKYLQGKENDSVALIQDSIKILEESGLGQSVACLRRLRYLAQVFLKSNRLSEAENVQRKILHIR EMSKV* |
| CDS seq | >XM_026531570.1 ATGTTTCTGCGACAATCTGGAAGTTTACTTCGAAGGAGATTAAGATACCTCCTCTCTACTAATACGATCACTACAACAAC TAAAACTACTATCAGTAGTCTCTCTTCATATCCTCCCCCAAGTAATGCGGGATTTCTCCACCACAATGGGCCTGAGATTC CAAGATGGCAGAAGCTTCACTGGGACTCTTCTTTGTGGATATTACTTTCTGGACAAGCTGCAATCATTCTTGGGGCAAGT GGAAATCTTGCATTTGCGGATGATGTTTCAGCTGAAGCGAGAACTGGTGCAGGGGAGGATGGGTCAAGTGATCTCCAGAG AGTTGAGGATGGGTCTGTGATATCAAATATCCACACATCTAAATGGCGGGTTTTTACAGATAAGGCGAGGGACTTTTTTT TGGAAGGAAAATTAGTGGAAGCTGAAAAACTATTTTTGTCAGCGGTGGAAGAAGCTAAAGAGGGTTTTGGGAAGAGCGAT CCACATGTTGCTGCTGCATTAAACAATTTGGCTGAGCTCTATAGAGTGAAGAAGGATTTCGCTAAAGCTGAGCCACTCTA TTTGGAGGCCGTCCATATTTTAGAGGAATCATTTGGACTTGGCGATATACGTGTTGCAGCCTCTCTACACAACCTTGGGC AGTTTTATATTGTGCAGAGGAAACTTGAAGAAGCCCATAACTGCTACGAGCGTGCGTTAAAGATAAAGAGGCGTGTTCTT GGACAAAACCACACAGACTATGCAAATACAATGTATCATCTGGGAACGGTGAAGTATCTTCAAGGAAAGGAGAATGATTC TGTGGCTCTAATTCAAGATTCTATTAAGATACTAGAGGAATCTGGCTTAGGACAATCAGTTGCTTGCTTGCGGAGGCTGC GGTATCTTGCTCAGGTATTTTTGAAGTCCAATCGACTTTCAGAGGCTGAGAATGTACAGAGAAAAATCTTGCACATCAGG GAGATGTCAAAGGTTTAA |
| Microexon DNA seq | CGTGCGTTAAAG |
| Microexon Amino Acid seq | RALK |
| Microexon-tag DNA Seq | TTTTATATTGTGCAGAGGAAACTTGAAGAAGCCCATAACTGCTACGAGCGTGCGTTAAAGATAAAGAGGCGTGTTCTTGGACAAAACCACACAGACTATGCAAATACA |
| Microexon-tag Amino Acid seq | FYIVQRKLEEAHNCYERALKIKRRVLGQNHTDYANT |
| Transcript ID | Ps.30160.1 |
| Gene ID | Ps.30160 |
| Gene Name | NA |
| Pfam domain motif | TPR_12 |
| Motif E-value | 5.5e-14 |
| Motif start | 205 |
| Motif end | 279 |
| Protein seq | >Ps.30160.1 MFLRQSGSLLRRRLRYLLSTNTITTTTKTTISSLSSYPPPSNAGFLHHNGPEIPRWQKLHWDSSLWILLSGQAAIILGAS GNLAFADDVSAEARTGAGEDGSSDLQRVEDGSVISNIHTSKWRVFTDKARDFFLEGKLVEAEKLFLSAVEEAKEGFGKSD PHVAAALNNLAELYRVKKDFAKAEPLYLEAVHILEESFGLGDIRVAASLHNLGQFYIVQRKLEEAHNCYERALKIKRRVL GQNHTDYANTMYHLGTVKYLQGKENDSVALIQDSIKILEVFLIIMFDINL* |
| CDS seq | >Ps.30160.1 ATGTTTCTGCGACAATCTGGAAGTTTACTTCGAAGGAGATTAAGATACCTCCTCTCTACTAATACGATCACTACAACAAC TAAAACTACTATCAGTAGTCTCTCTTCATATCCTCCCCCAAGTAATGCGGGATTTCTCCACCACAATGGGCCTGAGATTC CAAGATGGCAGAAGCTTCACTGGGACTCTTCTTTGTGGATATTACTTTCTGGACAAGCTGCAATCATTCTTGGGGCAAGT GGAAATCTTGCATTTGCGGATGATGTTTCAGCTGAAGCGAGAACTGGTGCAGGGGAGGATGGGTCAAGTGATCTCCAGAG AGTTGAGGATGGGTCTGTGATATCAAATATCCACACATCTAAATGGCGGGTTTTTACAGATAAGGCGAGGGACTTTTTTT TGGAAGGAAAATTAGTGGAAGCTGAAAAACTATTTTTGTCAGCGGTGGAAGAAGCTAAAGAGGGTTTTGGGAAGAGCGAT CCACATGTTGCTGCTGCATTAAACAATTTGGCTGAGCTCTATAGAGTGAAGAAGGATTTCGCTAAAGCTGAGCCACTCTA TTTGGAGGCCGTCCATATTTTAGAGGAATCATTTGGACTTGGCGATATACGTGTTGCAGCCTCTCTACACAACCTTGGGC AGTTTTATATTGTGCAGAGGAAACTTGAAGAAGCCCATAACTGCTACGAGCGTGCGTTAAAGATAAAGAGGCGTGTTCTT GGACAAAACCACACAGACTATGCAAATACAATGTATCATCTGGGAACGGTGAAGTATCTTCAAGGAAAGGAGAATGATTC TGTGGCTCTAATTCAAGATTCTATTAAGATACTAGAGGTTTTTCTGATCATCATGTTTGATATTAATCTGTAG |