
| Microexon ID | Pp_16:12744652-12744663:+ |
| Species | Physcomitrium patens | Coordinates | 16:12744652..12744663 |
| Microexon Cluster ID | MEP29 |
| Size | 12 |
| Phase | 0 |
| Pfam Domain Motif | TPR_12 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,12,48 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TTYTATCTTRTSCARMRSAARYTKGAWSAAGCYCRKRCWTGYTATGAGCGTGCTYTRAAGATAAARRGRCGTGTTYTGGGRSWTRRYCAYSCAGAYTATGCARAKACA |
| Logo of Microexon-tag DNA Seq | 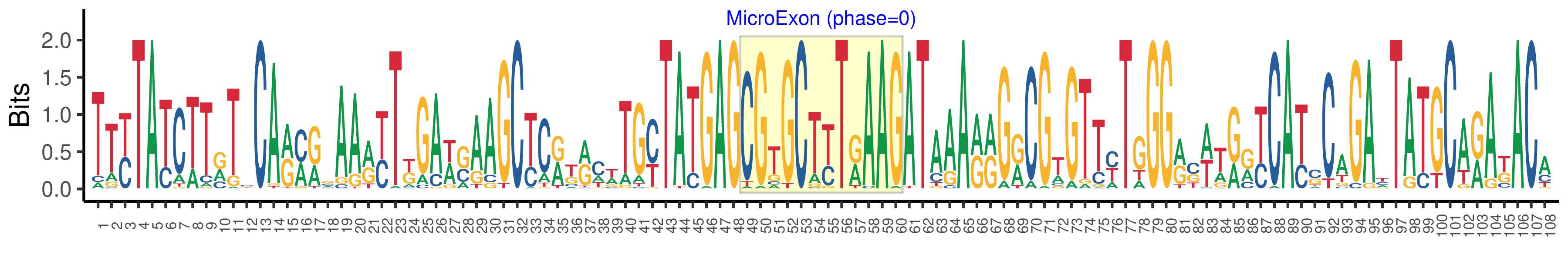 |
| Alignment of exons | 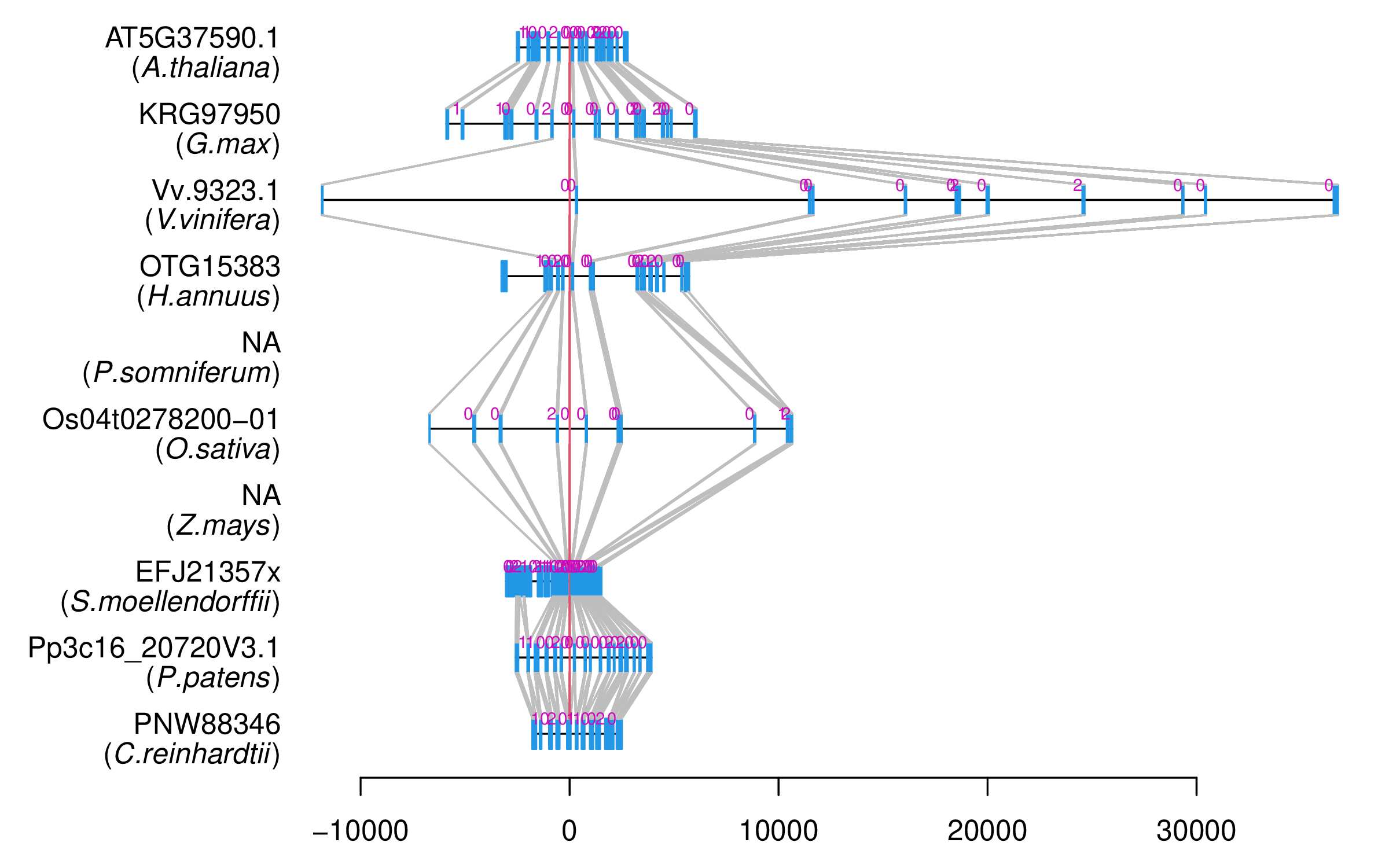 |
| Microexon DNA seq | CGCTCATTGAAG |
| Microexon Amino Acid seq | RSLK |
| Microexon-tag DNA Seq | ACTTACTTGCAGCAACACAATTATGAGCTGGCTCGGACATGCTATGAGCGCTCATTGAAGATTAAGGAAAGGAAGCTTGGACCTAATCATCCAGAGTACGCTAATACG |
| Microexon-tag Amino Acid Seq | TYLQQHNYELARTCYERSLKIKERKLGPNHPEYANT |
| Microexon-tag spanning region | 12744247-12744898 |
| Microexon-tag prediction score | 0.852 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | Pp3c16_20720V3.1x |
| Reference Transcript ID | Pp3c16_20720V3.1 |
| Gene ID | Pp3c16_20720 |
| Gene Name | NA |
| Transcript ID | Pp3c16_20720V3.1 |
| Protein ID | Pp3c16_20720V3.1 |
| Gene ID | Pp3c16_20720 |
| Gene Name | NA |
| Pfam domain motif | TPR_12 |
| Motif E-value | 1.7e-18 |
| Motif start | 206 |
| Motif end | 281 |
| Protein seq | >Pp3c16_20720V3.1 MGTRALGRLANSSHWNRSLASALLGSRESSFRSGKSLSSSYPVLGTTAGIFTSTDVVTNQSSFGRIWVYLISGVLSWGGI HTSLAEELSARPSTADNMEQEVQLVEDSLQSSNQASNPHTARWRIYTDMGRDLFSQGRLDEAEKYFVRALEEAKKGFGDR DPHVASSCNNLAELYRMRKEYAKAEPLFLESVQRLEQALGPMHESVAFALHNLAGTYLQQHNYELARTCYERSLKIKERK LGPNHPEYANTMFHLAEVLRLQGNNVDAEHLIRDSLRILEEGGVGHSQQAVRRMSRLAEILVHSGKLQEAEILQRKILHI LEMLQGPESLSTITAVDNLATTLQALRKLDEAEELFLRCLQVRQKALSSTHILVGSTLFKLAVVTTQQADEMVADKSRSV EEARMAYERAEDMLRQAIRIAEQRWEEALGGVAAADQAASAMLGTDVSLGPKALNTLIPPLLALVRSLNALGIVVMHLAD LAGSPVAAKERTQEAERVLRHCLTLLDKPPGIAQDLVEVPELQREYVSCLHHLAALLVRTKDQGSSENNKSAEEQAARLL SRAEGIEASHAARIQANLSN* |
| CDS seq | >Pp3c16_20720V3.1 ATGGGGACTAGGGCGCTTGGCAGATTGGCCAATTCTTCGCACTGGAATCGATCCCTTGCTTCTGCCTTGCTGGGATCGCG TGAATCGTCGTTTCGGTCGGGAAAGTCTCTGAGCTCGTCTTATCCTGTGCTAGGGACTACTGCTGGGATTTTCACCTCAA CAGACGTGGTCACAAATCAGTCATCTTTTGGAAGGATTTGGGTCTACTTGATCTCAGGAGTTCTTTCATGGGGTGGAATA CATACTAGTTTGGCTGAGGAACTAAGTGCACGACCATCAACTGCTGATAATATGGAACAGGAAGTTCAACTTGTTGAAGA CAGCCTACAATCTTCAAACCAAGCTTCCAATCCTCATACTGCAAGATGGAGGATCTACACTGACATGGGTCGCGACCTTT TCTCTCAGGGCCGTCTCGACGAGGCTGAGAAGTATTTTGTACGAGCTTTGGAGGAAGCCAAGAAAGGCTTTGGAGATCGA GATCCTCATGTTGCATCCAGCTGCAATAACCTGGCTGAGCTGTACCGCATGAGAAAGGAGTATGCCAAAGCAGAGCCTCT CTTTTTGGAGTCTGTGCAGAGGCTTGAACAAGCTTTGGGTCCTATGCATGAAAGTGTGGCGTTTGCGTTGCACAACCTTG CTGGGACTTACTTGCAGCAACACAATTATGAGCTGGCTCGGACATGCTATGAGCGCTCATTGAAGATTAAGGAAAGGAAG CTTGGACCTAATCATCCAGAGTACGCTAATACGATGTTTCATTTGGCAGAGGTACTGCGGCTTCAGGGCAATAACGTGGA TGCAGAGCATCTGATTCGTGATTCTTTGAGAATACTTGAGGAGGGTGGAGTTGGCCACTCGCAACAAGCTGTGCGACGTA TGAGCCGTCTTGCAGAAATACTGGTCCACTCTGGCAAGTTGCAAGAGGCAGAAATCCTCCAGCGCAAGATTCTTCATATT CTTGAAATGCTGCAGGGGCCTGAAAGTCTCAGCACAATCACAGCAGTTGATAATTTGGCCACAACCTTACAAGCCTTACG GAAACTGGATGAAGCTGAAGAACTCTTTCTGAGATGTCTCCAAGTTCGACAGAAGGCGCTTTCTTCAACACACATCCTGG TAGGATCAACATTGTTCAAATTAGCAGTGGTCACTACCCAGCAGGCCGACGAAATGGTGGCAGACAAAAGTCGTAGCGTG GAAGAAGCTCGAATGGCATATGAGAGGGCTGAGGACATGCTCCGTCAAGCAATTAGAATTGCAGAACAACGATGGGAAGA AGCGTTAGGAGGAGTTGCAGCTGCAGACCAAGCTGCATCTGCAATGTTGGGCACTGACGTCAGTTTAGGCCCTAAAGCTC TCAACACACTTATTCCACCTCTTCTGGCTCTTGTACGATCGTTGAATGCGCTTGGTATTGTAGTTATGCATCTTGCTGAT TTAGCTGGTTCCCCTGTGGCTGCAAAGGAAAGGACTCAAGAGGCTGAACGTGTTCTACGCCACTGCTTGACATTACTTGA CAAGCCACCTGGAATAGCACAGGACCTTGTTGAAGTTCCTGAGTTGCAGAGGGAATATGTGTCATGCCTCCATCATCTAG CGGCACTGCTTGTGAGGACAAAAGACCAGGGCTCAAGTGAAAATAACAAGTCAGCAGAGGAGCAAGCCGCAAGGCTGCTG AGTAGAGCAGAAGGTATTGAAGCTTCCCATGCAGCCCGAATACAAGCAAATCTTTCCAATTGA |
| Microexon DNA seq | CGCTCATTGAAG |
| Microexon Amino Acid seq | RSLK |
| Microexon-tag DNA Seq | ACTTACTTGCAGCAACACAATTATGAGCTGGCTCGGACATGCTATGAGCGCTCATTGAAGATTAAGGAAAGGAAGCTTGGACCTAATCATCCAGAGTACGCTAATACG |
| Microexon-tag Amino Acid seq | TYLQQHNYELARTCYERSLKIKERKLGPNHPEYANT |
| Transcript ID | Pp3c16_20720V3.2 |
| Gene ID | Pp.8155 |
| Gene Name | NA |
| Pfam domain motif | TPR_12 |
| Motif E-value | 1.3e-18 |
| Motif start | 206 |
| Motif end | 281 |
| Protein seq | >Pp3c16_20720V3.2 MGTRALGRLANSSHWNRSLASALLGSRESSFRSGKSLSSSYPVLGTTAGIFTSTDVVTNQSSFGRIWVYLISGVLSWGGI HTSLAEELSARPSTADNMEQEVQLVEDSLQSSNQASNPHTARWRIYTDMGRDLFSQGRLDEAEKYFVRALEEAKKGFGDR DPHVASSCNNLAELYRMRKEYAKAEPLFLESVQRLEQALGPMHESVAFALHNLAGTYLQQHNYELARTCYERSLKIKERK LGPNHPEYANTMFHLAEVLRLQGNNVDAEHLIRDSLRILEEGGVGHSQQAVRRMSRLAEILVHSGKLQEAEILQRKILHI LEMLQGPESLSTITAVDNLATTLQALRKLDEAEELFLRCLQVRQKALSSTHILVGSTLFKLAVVTTQQADEMVADKSRSV EEARMAYERAEDMLRQAIRIAEQRWEEALGGVAAADQAASAMLGTDVSLGPKALNTLIPPLLALVRSLNALGIVVMHLAD LAGSPVVRVCYLFHDLRL* |
| CDS seq | >Pp3c16_20720V3.2 ATGGGGACTAGGGCGCTTGGCAGATTGGCCAATTCTTCGCACTGGAATCGATCCCTTGCTTCTGCCTTGCTGGGATCGCG TGAATCGTCGTTTCGGTCGGGAAAGTCTCTGAGCTCGTCTTATCCTGTGCTAGGGACTACTGCTGGGATTTTCACCTCAA CAGACGTGGTCACAAATCAGTCATCTTTTGGAAGGATTTGGGTCTACTTGATCTCAGGAGTTCTTTCATGGGGTGGAATA CATACTAGTTTGGCTGAGGAACTAAGTGCACGACCATCAACTGCTGATAATATGGAACAGGAAGTTCAACTTGTTGAAGA CAGCCTACAATCTTCAAACCAAGCTTCCAATCCTCATACTGCAAGATGGAGGATCTACACTGACATGGGTCGCGACCTTT TCTCTCAGGGCCGTCTCGACGAGGCTGAGAAGTATTTTGTACGAGCTTTGGAGGAAGCCAAGAAAGGCTTTGGAGATCGA GATCCTCATGTTGCATCCAGCTGCAATAACCTGGCTGAGCTGTACCGCATGAGAAAGGAGTATGCCAAAGCAGAGCCTCT CTTTTTGGAGTCTGTGCAGAGGCTTGAACAAGCTTTGGGTCCTATGCATGAAAGTGTGGCGTTTGCGTTGCACAACCTTG CTGGGACTTACTTGCAGCAACACAATTATGAGCTGGCTCGGACATGCTATGAGCGCTCATTGAAGATTAAGGAAAGGAAG CTTGGACCTAATCATCCAGAGTACGCTAATACGATGTTTCATTTGGCAGAGGTACTGCGGCTTCAGGGCAATAACGTGGA TGCAGAGCATCTGATTCGTGATTCTTTGAGAATACTTGAGGAGGGTGGAGTTGGCCACTCGCAACAAGCTGTGCGACGTA TGAGCCGTCTTGCAGAAATACTGGTCCACTCTGGCAAGTTGCAAGAGGCAGAAATCCTCCAGCGCAAGATTCTTCATATT CTTGAAATGCTGCAGGGGCCTGAAAGTCTCAGCACAATCACAGCAGTTGATAATTTGGCCACAACCTTACAAGCCTTACG GAAACTGGATGAAGCTGAAGAACTCTTTCTGAGATGTCTCCAAGTTCGACAGAAGGCGCTTTCTTCAACACACATCCTGG TAGGATCAACATTGTTCAAATTAGCAGTGGTCACTACCCAGCAGGCCGACGAAATGGTGGCAGACAAAAGTCGTAGCGTG GAAGAAGCTCGAATGGCATATGAGAGGGCTGAGGACATGCTCCGTCAAGCAATTAGAATTGCAGAACAACGATGGGAAGA AGCGTTAGGAGGAGTTGCAGCTGCAGACCAAGCTGCATCTGCAATGTTGGGCACTGACGTCAGTTTAGGCCCTAAAGCTC TCAACACACTTATTCCACCTCTTCTGGCTCTTGTACGATCGTTGAATGCGCTTGGTATTGTAGTTATGCATCTTGCTGAT TTAGCTGGTTCCCCTGTGGTAAGAGTCTGTTATCTGTTCCATGATCTTAGATTATAG |