
| Microexon ID | Gm_11:30878240-30878251:- |
| Species | Glycine max | Coordinates | 11:30878240..30878251 |
| Microexon Cluster ID | MEP29 |
| Size | 12 |
| Phase | 0 |
| Pfam Domain Motif | TPR_12 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 48,12,48 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | TTYTATCTTRTSCARMRSAARYTKGAWSAAGCYCRKRCWTGYTATGAGCGTGCTYTRAAGATAAARRGRCGTGTTYTGGGRSWTRRYCAYSCAGAYTATGCARAKACA |
| Logo of Microexon-tag DNA Seq | 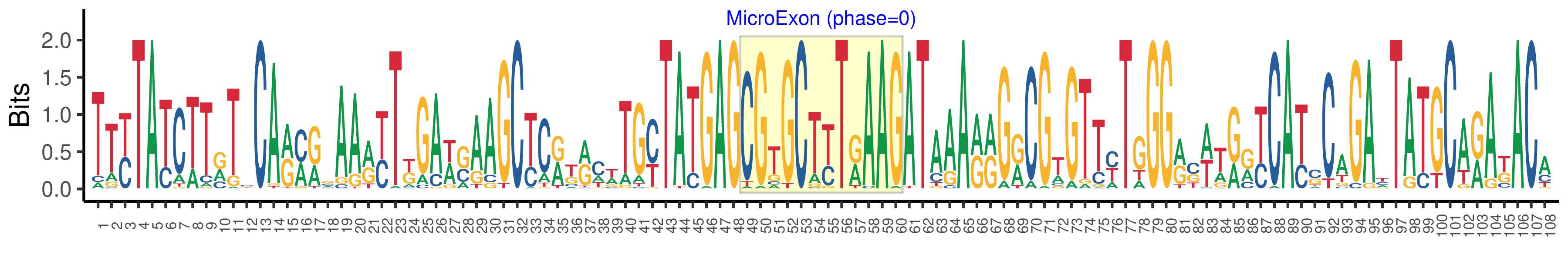 |
| Alignment of exons | 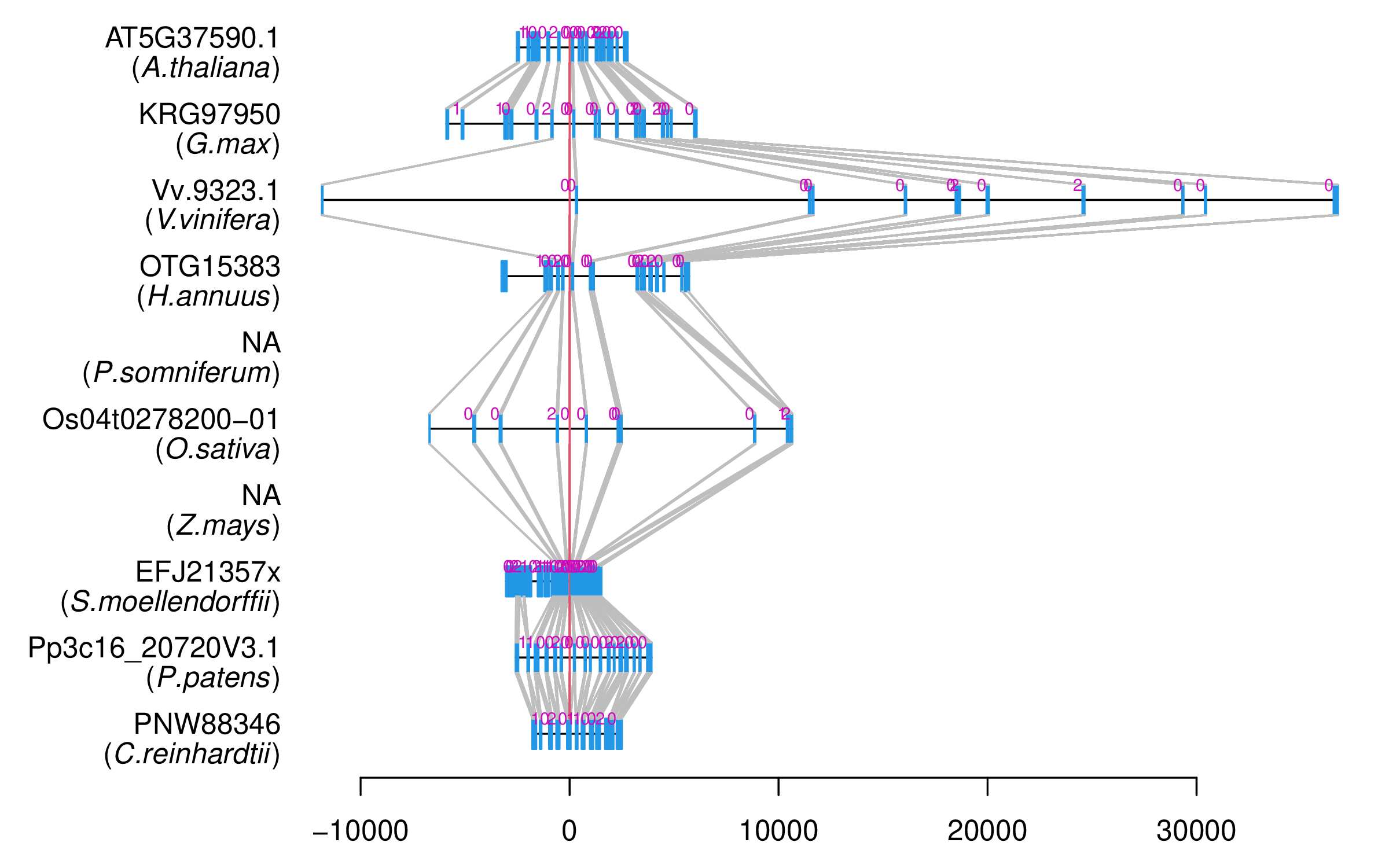 |
| Microexon DNA seq | CGTGCTTTGAAG |
| Microexon Amino Acid seq | RALK |
| Microexon-tag DNA Seq | TTCTACCTTGGACAGCGGAAGTTGGAAGAAGCTCGTGTTAGCTATGAGCGTGCTTTGAAGATAAAAAGGCGTGTTCTGGGGTATGGCCATTCAGAATGTTCAGATACA |
| Microexon-tag Amino Acid Seq | FYLGQRKLEEARVSYERALKIKRRVLGYGHSECSDT |
| Microexon-tag spanning region | 30878044-30879085 |
| Microexon-tag prediction score | 0.9674 |
| Overlapped with the annotated transcript (%) | 88.89 |
| New Transcript ID | KRH30928x |
| Reference Transcript ID | KRH30928 |
| Gene ID | GLYMA_11G215400 |
| Gene Name | NA |
Gm_11:30878240-30878251:- does not have available information here.
| Microexon DNA seq | CGTGCTTTGAAG |
| Microexon Amino Acid seq | RALK |
| Microexon-tag DNA Seq | TTCTACCTTGGACAGCGGAAGTTGGAAGAAGCTCGTGTTAGCTATGAGCGTGCTTTGAAGATAAAAAGGCGTGTTCTGGGGTATGGCCATTCAGAATGTTCAGATACA |
| Microexon-tag Amino Acid seq | FYLGQRKLEEARVSYERALKIKRRVLGYGHSECSDT |
| Transcript ID | Gm.7418.1 |
| Gene ID | Gm.7418 |
| Gene Name | NA |
| Pfam domain motif | TPR_12 |
| Motif E-value | 6.7e-12 |
| Motif start | 159 |
| Motif end | 231 |
| Protein seq | >Gm.7418.1 MSLRVAAASLLKKLRFPASHFFAASTATTNAFSGKLKLFRWYQTGAHGCRIDLRDPCLWIVMSGHVAMTLGISASTVFAE DATTEASSDNDPGGDLIGLRKIEDDSVVSNIHTAKWRVFTDKARQFFLQGKLDEAEKLFLSAIEEAKEGFGEKDPHVASA CNNLAELYRVKKAFDKAEPLYLEAINILEESFGPDDVRVGVAVHNLGQFYLGQRKLEEARVSYERALKIKRRVLGYGHSE CSDTMYHLGVVLYLQGKERDAEALIKDSIRMLEEGGEGESFVCIRRLRYLSQIYMKSHRIAEAEMVQRKILHVMELSKGW YFLDTVIAAESLALTLQASGNTKDSKELLERCLNVRKDLLPSDHIQIGANLLHLARVAMLDCSQHKKLDVSRAKAELDMA KNHLYNSIRISRQCLEKVLKQKDKLKKFSKPGDSRKEGQVALAILLQSLYTLSSVELDKQELQEIQERDNINIEAQEALL QCISTYKEFVHKRSIADTPEIKNEYLSCLKRAQNLFGHKVDE* |
| CDS seq | >Gm.7418.1 ATGTCTCTGCGTGTGGCGGCAGCAAGTTTGCTAAAAAAACTTCGCTTTCCGGCTTCACATTTCTTTGCTGCGTCTACAGC AACCACAAATGCATTTTCCGGTAAACTCAAACTTTTCCGTTGGTATCAAACTGGTGCACATGGTTGTAGGATAGATCTCA GGGATCCTTGTCTATGGATTGTTATGTCTGGACATGTAGCAATGACTTTAGGAATTAGTGCCAGCACTGTATTTGCAGAA GATGCAACAACTGAAGCATCTTCTGATAATGACCCAGGGGGTGACTTAATCGGATTGAGAAAAATTGAGGATGACTCTGT AGTATCAAACATACATACAGCAAAATGGAGAGTCTTTACAGATAAAGCGAGACAATTTTTTCTTCAGGGAAAACTAGATG AAGCTGAAAAACTCTTCCTTTCTGCTATAGAAGAAGCTAAAGAAGGTTTTGGCGAGAAAGACCCGCATGTTGCTTCTGCA TGCAACAATCTGGCAGAATTGTACAGGGTCAAAAAGGCATTTGACAAAGCAGAACCCTTGTATTTGGAAGCTATCAACAT ATTAGAGGAATCTTTTGGTCCTGATGATGTACGGGTTGGGGTTGCTGTTCACAACCTTGGGCAGTTCTACCTTGGACAGC GGAAGTTGGAAGAAGCTCGTGTTAGCTATGAGCGTGCTTTGAAGATAAAAAGGCGTGTTCTGGGGTATGGCCATTCAGAA TGTTCAGATACAATGTATCATCTAGGAGTGGTACTGTATCTTCAAGGAAAAGAAAGGGATGCTGAGGCCCTTATTAAGGA CTCTATAAGGATGCTAGAGGAAGGTGGTGAAGGAGAGTCTTTTGTATGCATTAGAAGACTACGATACCTTTCACAGATAT ATATGAAGTCACATCGAATTGCTGAAGCTGAAATGGTCCAGAGAAAGATCCTGCATGTTATGGAATTGTCAAAGGGATGG TATTTCTTGGACACTGTTATTGCAGCTGAATCTTTGGCTCTGACCCTGCAAGCATCTGGCAATACAAAAGATTCTAAAGA GCTTCTTGAAAGATGTCTTAATGTCCGGAAGGACTTACTTCCCAGTGACCATATTCAGATAGGTGCAAACTTACTTCATC TAGCAAGAGTGGCAATGCTTGATTGCAGTCAACATAAAAAGTTGGATGTTTCTAGAGCTAAAGCTGAGCTTGATATGGCA AAGAATCATTTGTATAATTCCATAAGGATTTCACGTCAATGTTTGGAGAAGGTATTGAAACAAAAGGACAAGTTAAAGAA ATTCAGTAAGCCTGGAGATTCTAGAAAGGAAGGCCAAGTAGCACTGGCCATATTGTTGCAATCACTCTATACGTTGTCAT CAGTGGAGTTAGATAAGCAAGAATTGCAGGAGATTCAGGAAAGAGATAATATTAACATCGAGGCTCAGGAGGCACTTCTT CAATGCATTTCTACTTACAAGGAGTTTGTACATAAGAGGTCAATTGCTGATACCCCTGAAATAAAGAACGAGTATCTTTC ATGTTTAAAGCGCGCTCAAAATTTGTTTGGTCATAAAGTTGATGAATAG |