
| Microexon ID | Sb_7:60038388-60038399:+ |
| Species | Sorghum Bicolor | Coordinates | 7:60038388..60038399 |
| Microexon Cluster ID | MEP28 |
| Size | 12 |
| Phase | 0 |
| Pfam Domain Motif | Peptidase_M1 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 24,19,5,12,48 |
| Microexon location in the Microexon-tag | 4 |
| Microexon-tag DNA Seq | GTTMGRCCWCAYTCTTAYATYAAGATGGACAACTTCTAYACAGTRACGGTKTATGARAAGGGWGCTGAAGTTGTCMGRATGTACAARACMTTRYTKGGRAGTYMAGGR |
| Logo of Microexon-tag DNA Seq | 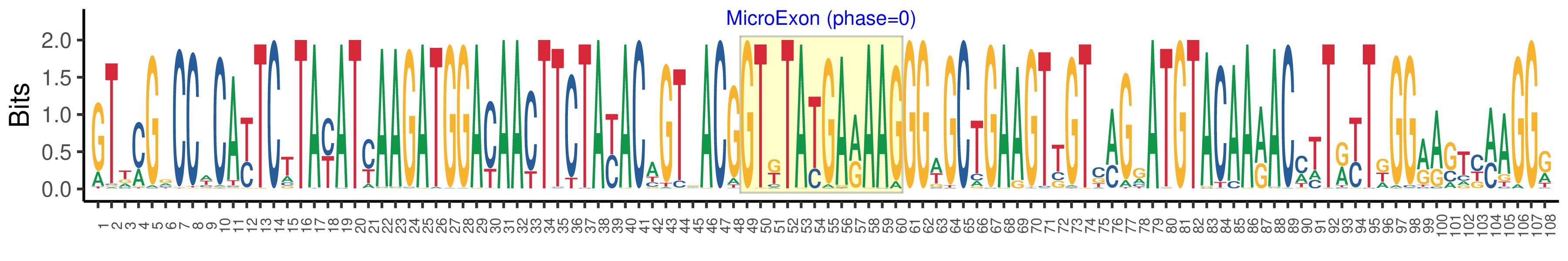 |
| Alignment of exons |  |
| Microexon DNA seq | GTTTACGAGAAG |
| Microexon Amino Acid seq | VYEK |
| Microexon-tag DNA Seq | ATCCGCCCTCATTCCTATATCAAGATGGACAACTTCTATACTGTGACGGTTTACGAGAAGGGTGCTGAAGTTGTCAGAATGTACAAGACCATGTTTGGAGCGTCAGGG |
| Microexon-tag Amino Acid Seq | IRPHSYIKMDNFYTVTVYEKGAEVVRMYKTMFGASG |
| Microexon-tag spanning region | 60037972-60038530 |
| Microexon-tag prediction score | 0.9703 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | EES14057x |
| Reference Transcript ID | EES14057 |
| Gene ID | SORBI_3007G165600 |
| Gene Name | NA |
| Transcript ID | EES14057 |
| Protein ID | |
| Gene ID | SORBI_3007G165600 |
| Gene Name | |
| Pfam domain motif | Peptidase_M1 |
| Motif E-value | 2.30E-50 |
| Motif start | 329 |
| Motif end | 540 |
| Protein seq | >EES14057 MARRLISPAAACRGCGFVRPGFLAAFSSFHYRTLPSCSPKRTSSVKSCFSNTYNLRKEGSRWIRSEPPLSLNRAKFVGKR TSCSVATEPPPAATEEPNMDAPKEIFLKDYKAPDYYFDTVDLQFQLGEEKTIVTSKIVVSPGVEAGNSAPLFLHGRDLKL LSIKVNGTELKGEEYKVNSRHLTLLTPPAGVFNLEIVTEIYPQLNTSLEGLYKSTGNFCTQCEAEGFRKITFFQDRPDVM AKYTCRIEADKTLYPVLLSNGNLIEQGDLEGGKHYALWEDPFKKPSYLFALVAGQLGCREDSFVTCSGRNVTLRIWTPAQ DLPKTAHAMYSLKAAMKWDEEVFGLEYDLDLFNIVVVPDFNMGAMENKSLNIFQSRLVLASPETATDGDYAAILGVVGHE YFHNWTGNRVTCRDWFQLTLKEGLTVFRDQEFSSDLGCRTVKRIADVSKLRIYQFPQDAGPMAHPIRPHSYIKMDNFYTV TVYEKGAEVVRMYKTMFGASGFRKGMDLYFQRHDGQAVTCEDFYAAMCDANNAQLPNFLQWYSQAGTPIVKVASSYDPSS QTFSLKFSQEVPPTPGQPVKEPMFIPVAVGLVDPTGKDMPLTSVYSDGTLQTLSTDGQPVFTTVLQFKKKEEEFIFKNIP ERPVPSLLRGYSAPVRLDSDLSESDLFFLLANDSDEFNRWEAGQVLARKLMLSLVADFQQQKTLALNPKFVDGIRSILRN TSLDKEFIAKAITLPGQGEIMDMMEVADPDAVHAVRNFIKKELALQLKDDLLAAVKSNRSSEAYTFDHDSMARRALKNTC LAYLASLNEPDFTELALHEYKSATNMTEQFAALAALSQNPGPVRDDALLDFYNKWQDDYLVVSKWFALQATSEIPGNVAN VQKLLSHPAFDMRNPNKVYSLIGGFCGSPVNFHAKDGSGYKFLGEIVLQLDKINPQVASRMVSAFSRWRRYDKTRQDLAK AQLEMIVSANGLSENVYEIASKSLAG* |
| CDS seq | >EES14057 ATGGCGCGTCGCCTGATTAGCCCCGCAGCGGCGTGCCGAGGCTGCGGTTTCGTTAGGCCCGGCTTTCTGGCTGCCTTCTC CTCCTTCCATTATCGAACCTTGCCATCGTGCTCACCGAAGAGGACTTCCTCAGTAAAGAGTTGCTTTAGCAACACTTACA ATTTAAGGAAAGAGGGTTCACGTTGGATAAGATCCGAGCCTCCCTTGTCACTAAATAGGGCAAAGTTTGTTGGCAAGAGG ACATCTTGTTCTGTTGCAACAGAGCCGCCTCCTGCTGCCACTGAAGAACCAAACATGGATGCCCCCAAGGAGATATTCTT GAAAGATTACAAGGCCCCTGATTATTATTTTGACACGGTGGATTTGCAATTTCAACTTGGTGAAGAGAAGACCATAGTTA CATCTAAAATAGTTGTATCTCCTGGAGTTGAAGCTGGCAACTCAGCTCCACTATTTCTTCATGGGCGTGACCTCAAGCTG TTATCAATCAAAGTCAATGGCACTGAATTAAAGGGTGAAGAGTATAAAGTCAATTCTCGCCATCTGACACTTTTAACACC ACCTGCTGGCGTATTCAACTTGGAAATAGTTACAGAAATATATCCTCAGCTAAACACATCATTGGAGGGGCTGTACAAGT CTACTGGTAACTTCTGCACTCAGTGTGAAGCAGAAGGGTTTCGGAAGATTACCTTCTTTCAGGATCGCCCGGATGTTATG GCAAAATATACTTGCCGTATTGAAGCTGATAAAACTTTGTATCCAGTATTATTGTCCAATGGGAATCTTATTGAGCAGGG AGACCTTGAGGGTGGAAAGCATTATGCACTGTGGGAGGATCCCTTCAAGAAACCAAGCTACCTGTTTGCTCTAGTTGCTG GTCAGCTGGGATGCCGGGAGGATTCATTTGTTACATGCTCTGGCCGCAACGTAACACTAAGAATTTGGACTCCTGCCCAA GATTTACCAAAAACAGCACATGCTATGTATTCTCTCAAAGCAGCAATGAAGTGGGATGAGGAGGTCTTTGGTCTGGAGTA TGATCTTGATTTATTCAACATTGTTGTTGTTCCCGATTTCAATATGGGAGCAATGGAAAACAAGAGCTTGAATATATTTC AATCTAGACTTGTGTTGGCATCACCAGAGACTGCCACTGATGGTGATTATGCTGCAATTTTGGGTGTTGTTGGACATGAG TATTTCCACAACTGGACTGGGAATAGAGTAACTTGCCGTGATTGGTTCCAGCTAACCCTGAAAGAAGGACTGACCGTTTT CAGGGATCAGGAGTTCTCTTCGGATCTTGGCTGTCGTACGGTAAAACGTATTGCTGATGTCTCCAAACTCAGAATCTATC AATTCCCACAGGATGCTGGGCCTATGGCACATCCTATCCGCCCTCATTCCTATATCAAGATGGACAACTTCTATACTGTG ACGGTTTACGAGAAGGGTGCTGAAGTTGTCAGAATGTACAAGACCATGTTTGGAGCGTCAGGGTTCCGAAAGGGCATGGA TCTTTATTTCCAAAGGCATGATGGGCAAGCTGTCACTTGTGAAGACTTCTATGCAGCCATGTGTGATGCAAATAATGCAC AACTGCCAAACTTTTTACAATGGTACTCTCAAGCAGGTACGCCTATAGTCAAGGTGGCATCTTCATATGATCCTAGTTCT CAGACATTCTCTTTGAAATTTAGCCAAGAAGTACCACCCACTCCTGGCCAACCAGTTAAGGAGCCTATGTTTATACCTGT TGCTGTTGGGCTTGTTGACCCTACTGGAAAAGATATGCCCCTCACTTCCGTTTACAGTGATGGAACTCTCCAAACACTCT CTACTGATGGCCAGCCAGTCTTCACTACTGTGCTGCAGTTTAAAAAGAAGGAAGAAGAGTTTATATTTAAGAACATTCCC GAGAGGCCAGTTCCTTCTTTGTTGAGGGGATATAGTGCTCCTGTCCGTCTAGATTCTGATCTCAGTGAGAGTGACCTATT TTTCTTGCTTGCCAATGACTCAGATGAATTTAACAGATGGGAAGCTGGCCAAGTTTTGGCACGTAAGTTGATGCTCAGTC TTGTAGCTGACTTCCAGCAACAGAAAACACTGGCCCTAAATCCAAAATTTGTTGATGGGATTAGATCAATTCTGCGCAAC ACAAGCTTGGATAAGGAATTCATTGCGAAAGCAATAACTTTGCCTGGTCAAGGGGAGATTATGGATATGATGGAAGTTGC AGATCCTGATGCAGTTCATGCTGTCCGTAACTTTATTAAGAAGGAGCTTGCCTTGCAACTCAAAGATGATCTTCTTGCAG CTGTGAAAAGCAACAGGAGTTCTGAGGCTTACACATTTGACCATGATAGTATGGCTCGCCGTGCATTGAAAAATACATGC CTTGCATACCTTGCATCATTGAATGAGCCAGATTTCACTGAGCTTGCACTGCATGAGTACAAGTCTGCGACTAACATGAC AGAGCAGTTTGCTGCATTAGCAGCATTATCCCAAAATCCAGGCCCAGTTCGTGATGATGCCCTCTTGGATTTCTATAACA AGTGGCAGGATGACTACTTGGTTGTAAGCAAGTGGTTTGCTCTTCAAGCTACTTCAGAGATTCCTGGAAATGTTGCAAAT GTTCAGAAGTTGCTGTCTCACCCTGCTTTCGACATGAGGAATCCCAACAAGGTTTACTCGCTAATCGGTGGATTCTGTGG CTCACCTGTGAACTTCCACGCGAAAGATGGATCTGGGTACAAGTTCTTGGGTGAAATTGTTTTGCAGCTTGACAAGATAA ACCCTCAGGTGGCCTCCCGCATGGTTTCAGCATTTTCTCGGTGGAGGCGATATGACAAGACCAGACAAGATCTTGCCAAG GCCCAGCTGGAGATGATCGTGTCGGCCAATGGACTCTCCGAAAACGTGTACGAGATTGCTTCAAAAAGCTTGGCGGGTTA G |
Sb_7:60038388-60038399:+ does not have available information here.