
| Microexon ID | Vv_11:2021619-2021629:- |
| Species | Vistis vinifera | Coordinates | 11:2021619..2021629 |
| Microexon Cluster ID | MEP26 |
| Size | 11 |
| Phase | 1 |
| Pfam Domain Motif | PRMT5 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,11,48 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | RRYGAYAARACCAGYGCYGAYTAYTACTTCGAYTCCTACTCYCAYTTYGGTATTCATGAAGAAATGYTGAARGATRYWGTGAGRACWAARACWTAYCAAAATGTTATY |
| Logo of Microexon-tag DNA Seq | 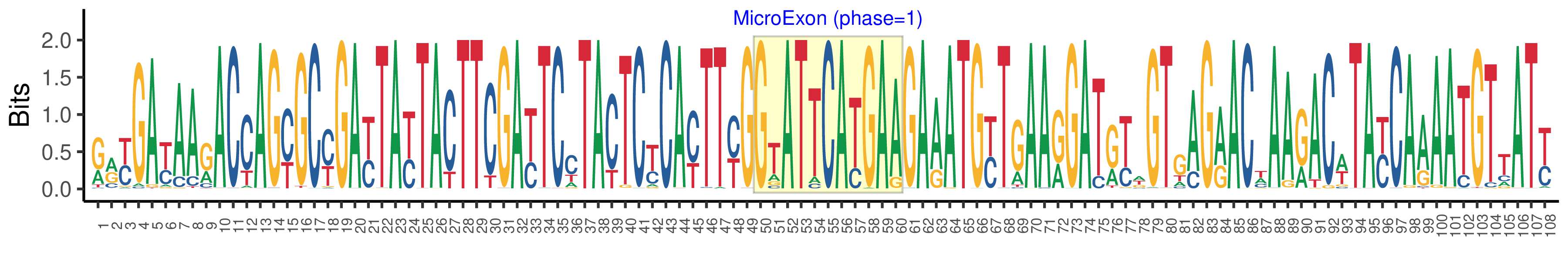 |
| Alignment of exons | 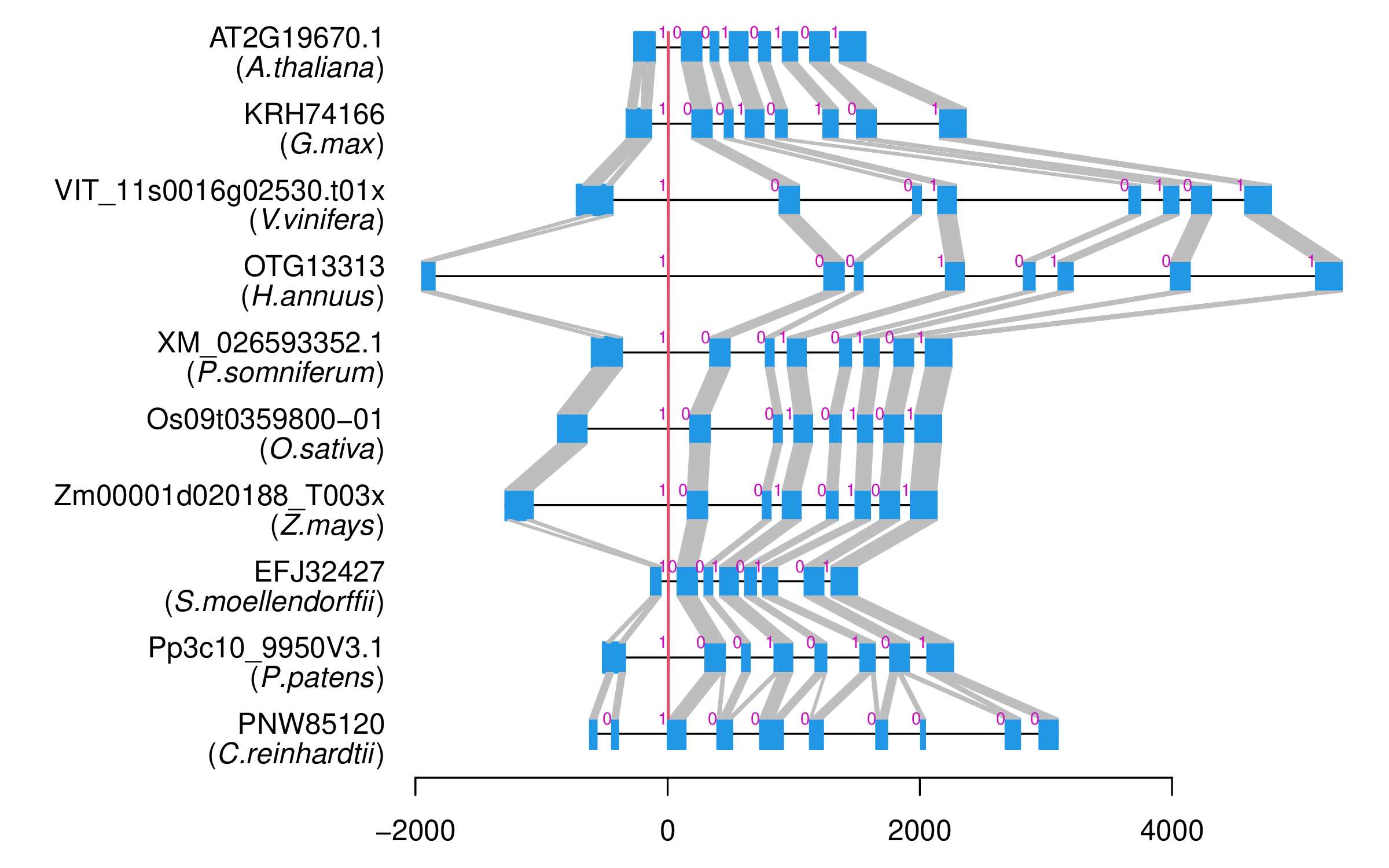 |
| Microexon DNA seq | GTATTCATGAA |
| Microexon Amino Acid seq | GIHE |
| Microexon-tag DNA Seq | AGTGATAAGACCAGCGCCGATTATTACTTCGACTCCTATTCTCACTTCGGTATTCATGAAGAAATGTTGAAGGATGTTGTGAGAACTAAGACCTATCAAAATGTCATT |
| Microexon-tag Amino Acid Seq | SDKTSADYYFDSYSHFGIHEEMLKDVVRTKTYQNVI |
| Microexon-tag spanning region | 2020695-2022113 |
| Microexon-tag prediction score | 0.9881 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | VIT_11s0016g02530.t01x |
| Reference Transcript ID | VIT_11s0016g02530.t01 |
| Gene ID | VIT_11s0016g02530 |
| Gene Name | NA |
| Transcript ID | VIT_11s0016g02530.t01 |
| Protein ID | VIT_11s0016g02530.t01 |
| Gene ID | VIT_11s0016g02530 |
| Gene Name | NA |
| Pfam domain motif | PRMT5 |
| Motif E-value | 0.0041 |
| Motif start | 44 |
| Motif end | 146 |
| Protein seq | >VIT_11s0016g02530.t01 MCEPGESDKDIARSFELDDSVIGSDKTSADYYFDSYSHFGIHEEMLKDVVRTKTYQNVIYKNKFLFKNKVVLDVGAGTGI LSLFCAKAGAKHVYAVECSHMADMAKEIVEVNGFSDVITVMKGKVEEIVLPVAQVDIIISEWMGYFLLFENMLNTVLYAR DKWLVNDGIVLPDKASLYLTAIEDAEYKEDKIEFWNSVYGFDMSCIKKQAMMEPLVDTVDQNQIVTNCQLLKTMDISKMA PGDASFTAPFKLVASRDDYIHALVAYFDVSFTKCHKLTGFSTGPRSRATHWKQTVLYLEDVLTICEGETVVGSMTVAQNK KNPRDVDIMIKYSFNGQRCQVSRTQYYKMR* |
| CDS seq | >VIT_11s0016g02530.t01 ATGTGCGAGCCTGGCGAGTCTGATAAAGACATTGCACGATCTTTCGAGCTCGACGACTCCGTCATTGGCAGTGATAAGAC CAGCGCCGATTATTACTTCGACTCCTATTCTCACTTCGGTATTCATGAAGAAATGTTGAAGGATGTTGTGAGAACTAAGA CCTATCAAAATGTCATTTATAAGAATAAGTTTCTGTTCAAGAACAAAGTAGTCCTTGATGTGGGAGCAGGGACTGGAATT CTGTCCCTCTTTTGTGCAAAGGCAGGGGCAAAACATGTTTATGCGGTTGAGTGTTCCCATATGGCTGACATGGCAAAAGA GATTGTTGAAGTAAATGGATTTTCTGATGTTATAACAGTTATGAAAGGAAAGGTTGAAGAAATTGTTCTTCCTGTTGCTC AAGTGGATATCATTATATCAGAATGGATGGGCTACTTTTTGTTGTTTGAGAATATGTTAAATACAGTCCTATATGCTCGT GATAAATGGCTTGTGAATGATGGTATTGTGCTGCCAGACAAAGCATCTCTCTATTTGACAGCCATTGAGGATGCGGAGTA CAAAGAAGACAAGATTGAATTTTGGAACAGTGTGTATGGCTTTGACATGAGCTGTATCAAGAAGCAAGCAATGATGGAAC CACTTGTGGACACAGTTGATCAGAATCAGATCGTCACAAACTGCCAGTTACTCAAGACTATGGACATCTCTAAAATGGCT CCTGGGGATGCTTCCTTCACAGCTCCTTTTAAGCTTGTGGCATCACGCGATGATTACATCCATGCTCTTGTAGCATATTT TGATGTATCATTTACCAAGTGTCATAAATTGACAGGCTTCTCCACAGGGCCAAGATCACGGGCTACACATTGGAAACAAA CAGTCCTGTATCTGGAAGATGTGCTAACCATATGCGAAGGGGAGACAGTTGTTGGGAGCATGACAGTGGCCCAGAACAAG AAGAATCCTCGTGACGTTGATATAATGATCAAATATTCCTTCAATGGTCAGCGTTGCCAGGTCTCAAGAACTCAATATTA CAAGATGCGTTAG |
| Microexon DNA seq | GTATTCATGAA |
| Microexon Amino Acid seq | GIHE |
| Microexon-tag DNA Seq | AGTGATAAGACCAGCGCCGATTATTACTTCGACTCCTATTCTCACTTCGGTATTCATGAAGAAATGTTGAAGGATGTTGTGAGAACTAAGACCTATCAAAATGTCATT |
| Microexon-tag Amino Acid seq | SDKTSADYYFDSYSHFGIHEEMLKDVVRTKTYQNVI |
| Transcript ID | Vv.3356.1 |
| Gene ID | Vv.3356 |
| Gene Name | NA |
| Pfam domain motif | PRMT5 |
| Motif E-value | 0.0025 |
| Motif start | 100 |
| Motif end | 202 |
| Protein seq | >Vv.3356.1 MARRESHSHHPHHHQGTHTRFEDEQIEEATESSNLEDPMCDADESTIGQDKAIDDTMCEPGESDKDIARSFELDDSVIGS DKTSADYYFDSYSHFGIHEEMLKDVVRTKTYQNVIYKNKFLFKNKVVLDVGAGTGILSLFCAKAGAKHVYAVECSHMADM AKEIVEVNGFSDVITVMKGKVEEIVLPVAQVDIIISEWMGYFLLFENMLNTVLYARDKWLVNDGIVLPDKASLYLTAIED AEYKEDKIECEPIEIFKIHLSRIFFSFCLCFYVWER* |
| CDS seq | >Vv.3356.1 ATGGCTCGACGAGAGTCCCACAGCCACCACCCCCACCACCACCAAGGCACGCACACCAGATTCGAGGACGAACAGATCGA AGAAGCGACGGAGAGCTCCAACCTCGAAGATCCTATGTGCGACGCAGACGAGTCCACCATTGGCCAAGACAAAGCCATTG ATGACACGATGTGCGAGCCTGGCGAGTCTGATAAAGACATTGCACGATCTTTCGAGCTCGACGACTCCGTCATTGGCAGT GATAAGACCAGCGCCGATTATTACTTCGACTCCTATTCTCACTTCGGTATTCATGAAGAAATGTTGAAGGATGTTGTGAG AACTAAGACCTATCAAAATGTCATTTATAAGAATAAGTTTCTGTTCAAGAACAAAGTAGTCCTTGATGTGGGAGCAGGGA CTGGAATTCTGTCCCTCTTTTGTGCAAAGGCAGGGGCAAAACATGTTTATGCGGTTGAGTGTTCCCATATGGCTGACATG GCAAAAGAGATTGTTGAAGTAAATGGATTTTCTGATGTTATAACAGTTATGAAAGGAAAGGTTGAAGAAATTGTTCTTCC TGTTGCTCAAGTGGATATCATTATATCAGAATGGATGGGCTACTTTTTGTTGTTTGAGAATATGTTAAATACAGTCCTAT ATGCTCGTGATAAATGGCTTGTGAATGATGGTATTGTGCTGCCAGACAAAGCATCTCTCTATTTGACAGCCATTGAGGAT GCGGAGTACAAAGAAGACAAGATTGAATGTGAGCCTATTGAGATTTTTAAAATACATTTGAGTAGAATATTTTTCTCATT CTGTCTATGTTTTTATGTGTGGGAAAGATGA |