
| Microexon ID | Ps_NC_039359.1:146986436-146986446:- |
| Species | Papaver somniferum | Coordinates | NC_039359.1:146986436..146986446 |
| Microexon Cluster ID | MEP26 |
| Size | 11 |
| Phase | 1 |
| Pfam Domain Motif | PRMT5 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,11,48 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | RRYGAYAARACCAGYGCYGAYTAYTACTTCGAYTCCTACTCYCAYTTYGGTATTCATGAAGAAATGYTGAARGATRYWGTGAGRACWAARACWTAYCAAAATGTTATY |
| Logo of Microexon-tag DNA Seq | 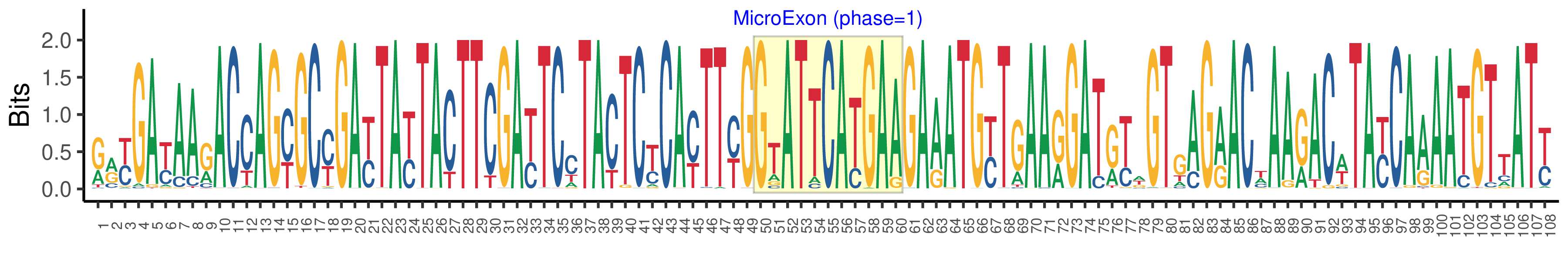 |
| Alignment of exons | 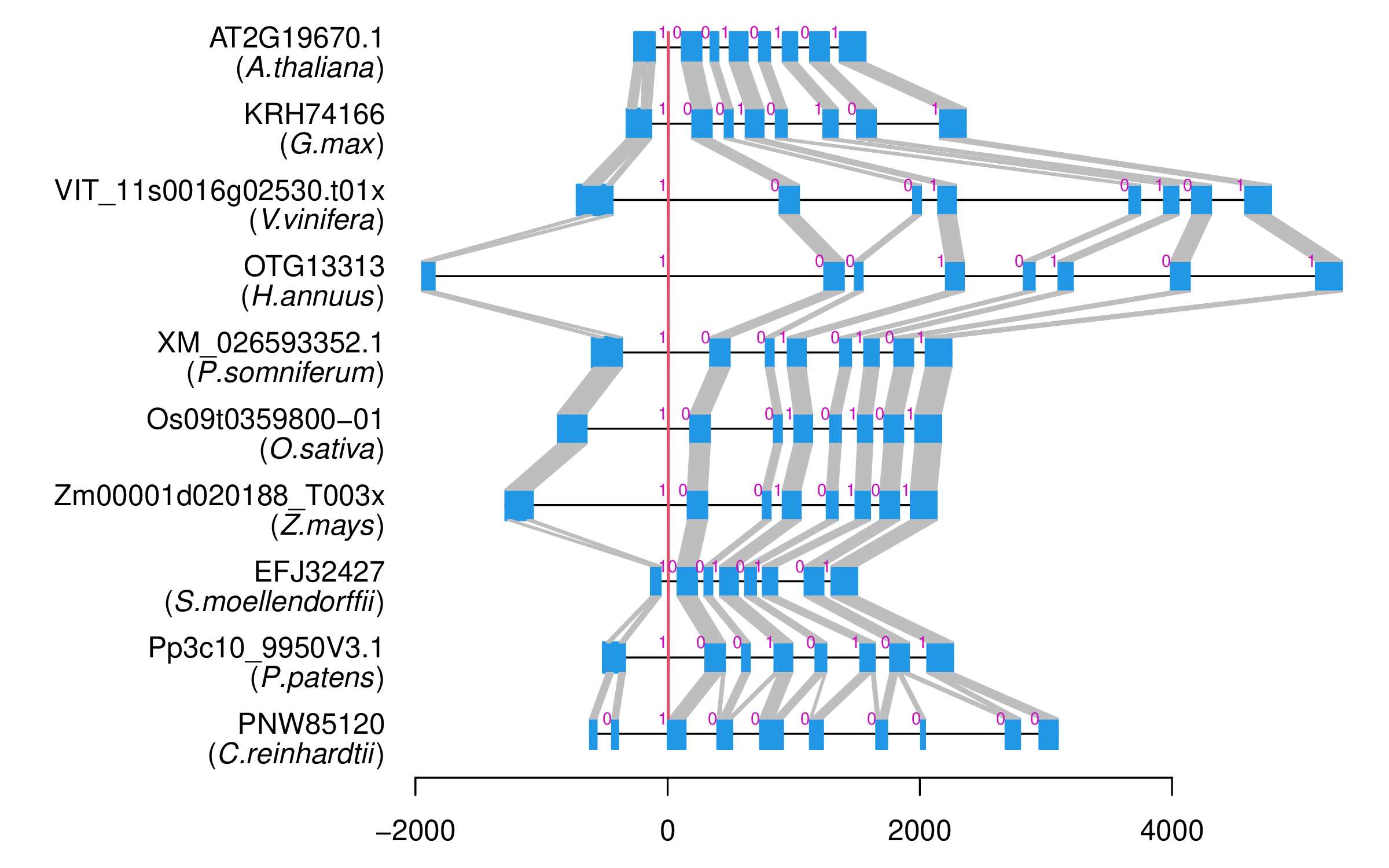 |
| Microexon DNA seq | GTATTCATGAA |
| Microexon Amino Acid seq | GIHE |
| Microexon-tag DNA Seq | GATGATAAAACTAGTGCTGATTATTACTTCGATTCTTATTCTCACTTTGGTATTCATGAAGAAATGTTGAAGGACACAGTGAGGACAAAGACATACCAAAATGTTATC |
| Microexon-tag Amino Acid Seq | DDKTSADYYFDSYSHFGIHEEMLKDTVRTKTYQNVI |
| Microexon-tag spanning region | 146986062-146986856 |
| Microexon-tag prediction score | 0.9768 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | XM_026593352.1x |
| Reference Transcript ID | XM_026593352.1 |
| Gene ID | NA |
| Gene Name | NA |
| Transcript ID | XM_026593352.1 |
| Protein ID | XP_026449137.1 |
| Gene ID | LOC113349384 |
| Gene Name | NA |
| Pfam domain motif | PRMT5 |
| Motif E-value | 4e-04 |
| Motif start | 85 |
| Motif end | 187 |
| Protein seq | >XP_026449137.1 MGRRRSNNGGSSSSSNIENNNHPQVTKLCIDDDDNELPEEMITPTTDQSMSEADNSTAVVGGGDDDKTSADYYFDSYSHF GIHEEMLKDTVRTKTYQNVIYKNTFLFKDKIVLDVGAGTGILSLFCAKAGAKHVYAIECSQMADMAKQIVETNGFSNVIT VLKGKVEEIELPVPKVDIIISEWMGYFLLFENMLDTVLYARNKWLVNDGIVLPDKASLYVTAIEDHEYKEDKIEFWNDVY GFDMKCIKKQSIMEPLVDTVDQNQIVTNYQLLKTMDILKMASGDVSFTAPFKLRAERSDYIHALVAYFDVSFTQCHKLMG FSTGPRSRTTHWKQTVLYLEDVLTICEGEVLSGSMTVSQNKKNPRDVDITLKYELNGQRSKISGTQHYKMR* |
| CDS seq | >XM_026593352.1 ATGGGCCGTAGAAGAAGCAACAATGGGGGTTCTTCATCATCAAGTAATATTGAGAATAATAATCATCCGCAAGTAACTAA GTTATGTATCGATGATGATGATAATGAACTACCAGAAGAGATGATTACTCCTACTACTGATCAATCTATGTCTGAAGCTG ATAATAGTACGGCTGTTGTTGGTGGTGGTGATGATGATAAAACTAGTGCTGATTATTACTTCGATTCTTATTCTCACTTT GGTATTCATGAAGAAATGTTGAAGGACACAGTGAGGACAAAGACATACCAAAATGTTATCTACAAGAACACGTTCCTCTT CAAGGATAAAATTGTTCTTGATGTTGGTGCTGGTACTGGAATTTTATCCCTCTTTTGTGCAAAAGCAGGAGCAAAGCATG TTTATGCGATCGAGTGTTCCCAAATGGCTGACATGGCGAAACAGATTGTTGAAACAAATGGCTTCTCTAATGTTATTACA GTTTTGAAGGGGAAGGTGGAAGAGATTGAGCTTCCAGTCCCCAAAGTTGATATCATCATATCCGAGTGGATGGGTTATTT TCTATTGTTTGAGAATATGTTAGACACTGTTCTTTATGCACGTAACAAGTGGCTTGTAAACGATGGGATTGTGTTGCCCG ACAAGGCGTCACTCTACGTAACAGCCATTGAGGATCATGAGTACAAGGAAGACAAAATTGAATTCTGGAATGATGTTTAC GGCTTCGACATGAAATGTATCAAAAAACAATCCATAATGGAACCCCTTGTAGACACGGTTGATCAAAATCAAATTGTCAC AAACTATCAGCTGCTGAAGACAATGGATATCTTGAAGATGGCTTCTGGAGATGTCTCATTTACAGCTCCCTTTAAGCTCA GGGCTGAACGTAGTGACTATATTCATGCCCTAGTTGCATACTTTGATGTCTCATTTACGCAGTGTCATAAGTTGATGGGC TTCTCTACAGGACCTAGATCCCGCACCACGCATTGGAAGCAAACGGTATTGTACCTTGAGGACGTGCTAACCATTTGCGA AGGTGAAGTACTGTCTGGGAGCATGACAGTTTCACAAAACAAGAAGAATCCTCGTGATGTTGATATTACTCTTAAATATG AATTGAATGGTCAGCGTAGCAAGATATCAGGAACTCAACATTACAAGATGCGTTGA |
| Microexon DNA seq | GTATTCATGAA |
| Microexon Amino Acid seq | GIHE |
| Microexon-tag DNA Seq | GATGATAAAACTAGTGCTGATTATTACTTCGATTCTTATTCTCACTTTGGTATTCATGAAGAAATGTTGAAGGACACAGTGAGGACAAAGACATACCAAAATGTTATC |
| Microexon-tag Amino Acid seq | DDKTSADYYFDSYSHFGIHEEMLKDTVRTKTYQNVI |
| Transcript ID | XM_026593352.1 |
| Gene ID | Ps.13044 |
| Gene Name | NA |
| Pfam domain motif | PRMT5 |
| Motif E-value | 4e-04 |
| Motif start | 85 |
| Motif end | 187 |
| Protein seq | >XM_026593352.1 MGRRRSNNGGSSSSSNIENNNHPQVTKLCIDDDDNELPEEMITPTTDQSMSEADNSTAVVGGGDDDKTSADYYFDSYSHF GIHEEMLKDTVRTKTYQNVIYKNTFLFKDKIVLDVGAGTGILSLFCAKAGAKHVYAIECSQMADMAKQIVETNGFSNVIT VLKGKVEEIELPVPKVDIIISEWMGYFLLFENMLDTVLYARNKWLVNDGIVLPDKASLYVTAIEDHEYKEDKIEFWNDVY GFDMKCIKKQSIMEPLVDTVDQNQIVTNYQLLKTMDILKMASGDVSFTAPFKLRAERSDYIHALVAYFDVSFTQCHKLMG FSTGPRSRTTHWKQTVLYLEDVLTICEGEVLSGSMTVSQNKKNPRDVDITLKYELNGQRSKISGTQHYKMR* |
| CDS seq | >XM_026593352.1 ATGGGCCGTAGAAGAAGCAACAATGGGGGTTCTTCATCATCAAGTAATATTGAGAATAATAATCATCCGCAAGTAACTAA GTTATGTATCGATGATGATGATAATGAACTACCAGAAGAGATGATTACTCCTACTACTGATCAATCTATGTCTGAAGCTG ATAATAGTACGGCTGTTGTTGGTGGTGGTGATGATGATAAAACTAGTGCTGATTATTACTTCGATTCTTATTCTCACTTT GGTATTCATGAAGAAATGTTGAAGGACACAGTGAGGACAAAGACATACCAAAATGTTATCTACAAGAACACGTTCCTCTT CAAGGATAAAATTGTTCTTGATGTTGGTGCTGGTACTGGAATTTTATCCCTCTTTTGTGCAAAAGCAGGAGCAAAGCATG TTTATGCGATCGAGTGTTCCCAAATGGCTGACATGGCGAAACAGATTGTTGAAACAAATGGCTTCTCTAATGTTATTACA GTTTTGAAGGGGAAGGTGGAAGAGATTGAGCTTCCAGTCCCCAAAGTTGATATCATCATATCCGAGTGGATGGGTTATTT TCTATTGTTTGAGAATATGTTAGACACTGTTCTTTATGCACGTAACAAGTGGCTTGTAAACGATGGGATTGTGTTGCCCG ACAAGGCGTCACTCTACGTAACAGCCATTGAGGATCATGAGTACAAGGAAGACAAAATTGAATTCTGGAATGATGTTTAC GGCTTCGACATGAAATGTATCAAAAAACAATCCATAATGGAACCCCTTGTAGACACGGTTGATCAAAATCAAATTGTCAC AAACTATCAGCTGCTGAAGACAATGGATATCTTGAAGATGGCTTCTGGAGATGTCTCATTTACAGCTCCCTTTAAGCTCA GGGCTGAACGTAGTGACTATATTCATGCCCTAGTTGCATACTTTGATGTCTCATTTACGCAGTGTCATAAGTTGATGGGC TTCTCTACAGGACCTAGATCCCGCACCACGCATTGGAAGCAAACGGTATTGTACCTTGAGGACGTGCTAACCATTTGCGA AGGTGAAGTACTGTCTGGGAGCATGACAGTTTCACAAAACAAGAAGAATCCTCGTGATGTTGATATTACTCTTAAATATG AATTGAATGGTCAGCGTAGCAAGATATCAGGAACTCAACATTACAAGATGCGTTGA |