
| Microexon ID | Ha_3:3367227-3367237:+ |
| Species | Helianthus annuus | Coordinates | 3:3367227..3367237 |
| Microexon Cluster ID | MEP26 |
| Size | 11 |
| Phase | 1 |
| Pfam Domain Motif | PRMT5 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,11,48 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | RRYGAYAARACCAGYGCYGAYTAYTACTTCGAYTCCTACTCYCAYTTYGGTATTCATGAAGAAATGYTGAARGATRYWGTGAGRACWAARACWTAYCAAAATGTTATY |
| Logo of Microexon-tag DNA Seq | 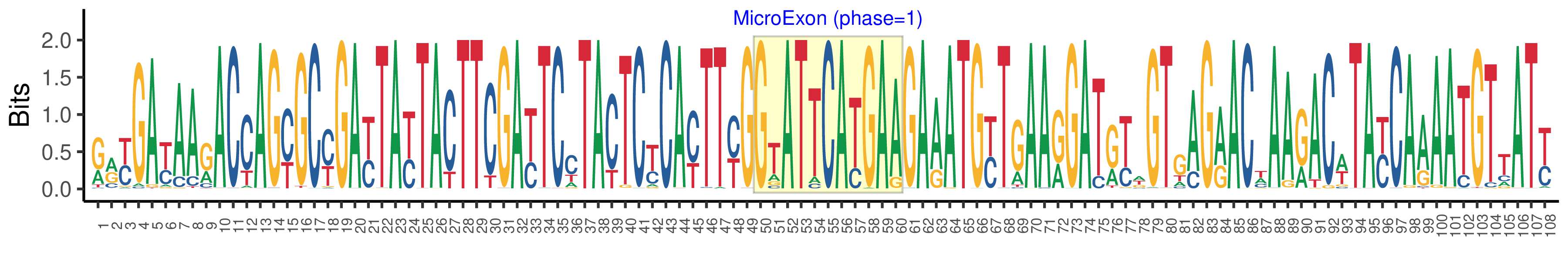 |
| Alignment of exons | 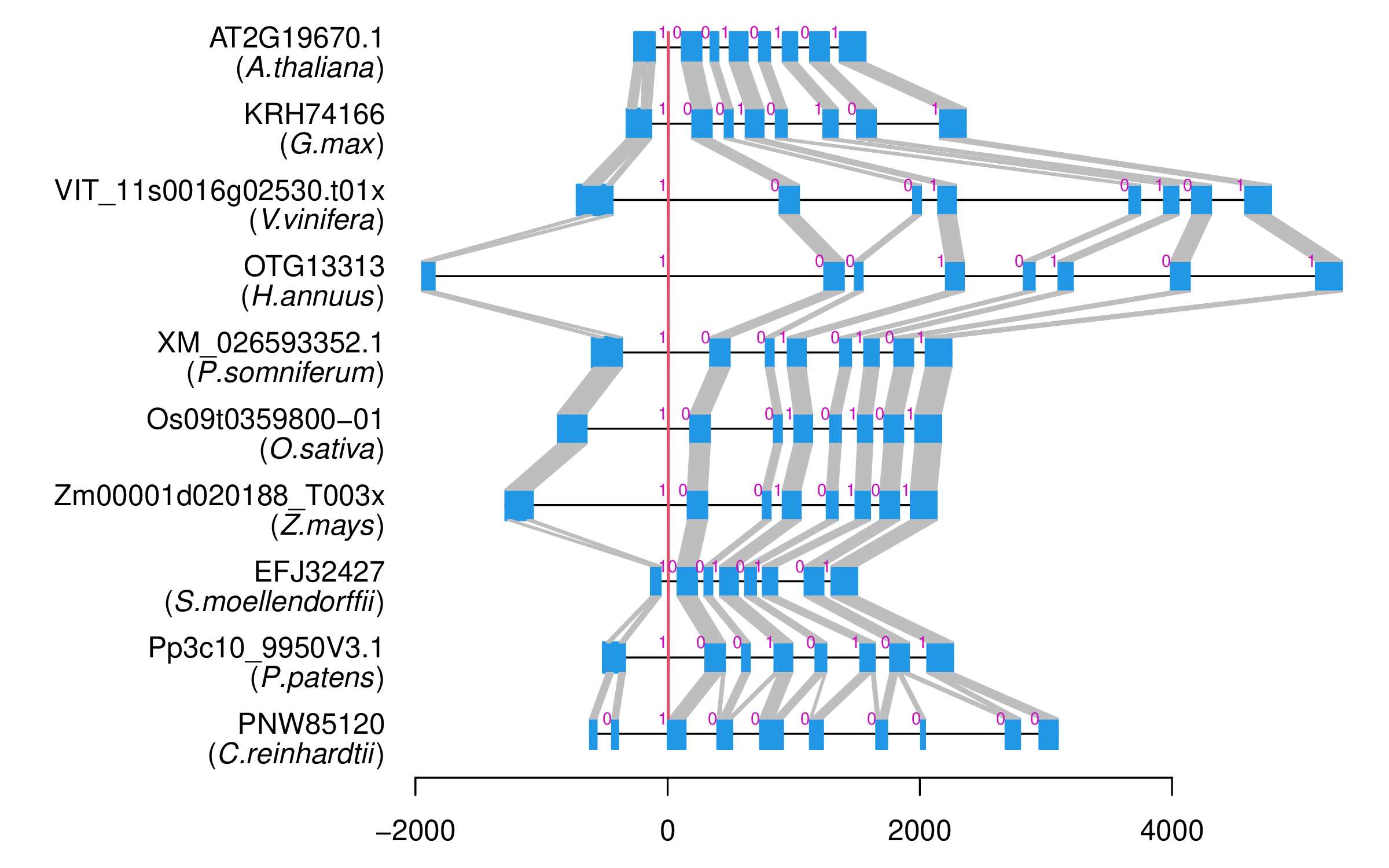 |
| Microexon DNA seq | GTATTCATGAG |
| Microexon Amino Acid seq | GIHE |
| Microexon-tag DNA Seq | ACTGATAAGACCAGTGCTGATTATTACTTCGATTCATACTCTCACTTTGGTATTCATGAGGAAATGCTGAAGGATGTGGTGAGAACCAAGACTTATCAAAATGTCATC |
| Microexon-tag Amino Acid Seq | TDKTSADYYFDSYSHFGIHEEMLKDVVRTKTYQNVI |
| Microexon-tag spanning region | 3366071-3367429 |
| Microexon-tag prediction score | 0.9835 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | OTG30082x |
| Reference Transcript ID | OTG30082 |
| Gene ID | HannXRQ_Chr03g0060541 |
| Gene Name | ANM1 |
| Transcript ID | OTG30082 |
| Protein ID | OTG30082 |
| Gene ID | HannXRQ_Chr03g0060541 |
| Gene Name | ANM1 |
| Pfam domain motif | PRMT5 |
| Motif E-value | 0.00078 |
| Motif start | 61 |
| Motif end | 163 |
| Protein seq | >OTG30082 MTDAMPPSKFEDADETIEEAATATSNMEEDSAMCEPHDSSTDKTSADYYFDSYSHFGIHEEMLKDVVRTKTYQNVIYKNK FLFKDKIVLDVGAGTGILSLFCAKAGAKHVYAIECSQMADMAQEIVKANGFSNVITVLKGKVEEIDLPVPQVDIIISEWM GYFLLYENMLNTVLYARDKWLVSDGIVLPDNATLHLTAIEDSEYKEDKIEFWKNVYGFDMSCIRKQSMMEPLVDTVDQNQ IVTNCQLLKSMNISKMTSGDASFTAPFKLVAERDDYIHALVAYFDVAFTVCHKMTGFSTGPRSKNTHWKQTVLYLEDVLT ICQGEFVVGTMTVTQNKKNPRDIDITLKYSLNGRRCNVSRTQEYKMR* |
| CDS seq | >OTG30082 ATGACTGACGCTATGCCGCCGTCAAAGTTCGAAGACGCCGATGAAACCATCGAAGAAGCAGCCACTGCAACCTCCAACAT GGAGGAAGATTCCGCCATGTGCGAACCTCACGATTCTTCCACTGATAAGACCAGTGCTGATTATTACTTCGATTCATACT CTCACTTTGGTATTCATGAGGAAATGCTGAAGGATGTGGTGAGAACCAAGACTTATCAAAATGTCATCTACAAAAACAAA TTTTTATTCAAAGACAAGATTGTTCTTGACGTTGGTGCCGGGACCGGAATATTATCCCTCTTCTGTGCAAAAGCTGGGGC AAAACATGTTTATGCTATTGAGTGCTCACAGATGGCTGACATGGCACAAGAGATTGTTAAGGCGAACGGTTTTTCAAATG TTATAACTGTTTTGAAGGGAAAAGTTGAAGAAATCGACCTTCCAGTTCCACAAGTCGATATTATTATTTCGGAATGGATG GGTTACTTTCTGTTATACGAGAATATGTTGAATACAGTTTTATATGCTCGTGATAAGTGGCTGGTTAGCGATGGAATCGT GCTACCTGATAATGCAACACTTCATCTGACAGCAATAGAGGACTCCGAATACAAAGAGGACAAGATTGAATTTTGGAAAA ACGTGTACGGCTTTGACATGAGCTGCATCAGGAAACAGTCGATGATGGAACCTCTTGTTGACACAGTTGACCAGAATCAG ATCGTTACAAATTGTCAGTTACTTAAGAGCATGAATATCTCCAAGATGACGTCTGGAGATGCATCGTTTACAGCGCCTTT CAAGCTTGTGGCCGAGCGTGATGATTACATTCATGCTCTTGTAGCTTACTTTGATGTCGCTTTTACTGTTTGCCACAAAA TGACAGGATTCTCTACAGGGCCAAGGTCAAAGAATACACATTGGAAACAAACAGTGCTATATCTCGAAGATGTGTTAACT ATATGCCAAGGGGAGTTTGTGGTTGGAACCATGACAGTGACGCAAAACAAGAAAAATCCCCGAGACATTGACATCACACT CAAATACTCATTAAACGGTCGTCGTTGCAATGTGTCGAGGACCCAAGAATACAAGATGCGTTGA |
| Microexon DNA seq | GTATTCATGAG |
| Microexon Amino Acid seq | GIHE |
| Microexon-tag DNA Seq | ACTGATAAGACCAGTGCTGATTATTACTTCGATTCATACTCTCACTTTGGTATTCATGAGGAAATGCTGAAGGATGTGGTGAGAACCAAGACTTATCAAAATGTCATC |
| Microexon-tag Amino Acid seq | TDKTSADYYFDSYSHFGIHEEMLKDVVRTKTYQNVI |
| Transcript ID | OTG30082 |
| Gene ID | Ha.36162 |
| Gene Name | ANM1 |
| Pfam domain motif | PRMT5 |
| Motif E-value | 0.00078 |
| Motif start | 61 |
| Motif end | 163 |
| Protein seq | >OTG30082 MTDAMPPSKFEDADETIEEAATATSNMEEDSAMCEPHDSSTDKTSADYYFDSYSHFGIHEEMLKDVVRTKTYQNVIYKNK FLFKDKIVLDVGAGTGILSLFCAKAGAKHVYAIECSQMADMAQEIVKANGFSNVITVLKGKVEEIDLPVPQVDIIISEWM GYFLLYENMLNTVLYARDKWLVSDGIVLPDNATLHLTAIEDSEYKEDKIEFWKNVYGFDMSCIRKQSMMEPLVDTVDQNQ IVTNCQLLKSMNISKMTSGDASFTAPFKLVAERDDYIHALVAYFDVAFTVCHKMTGFSTGPRSKNTHWKQTVLYLEDVLT ICQGEFVVGTMTVTQNKKNPRDIDITLKYSLNGRRCNVSRTQEYKMR* |
| CDS seq | >OTG30082 ATGACTGACGCTATGCCGCCGTCAAAGTTCGAAGACGCCGATGAAACCATCGAAGAAGCAGCCACTGCAACCTCCAACAT GGAGGAAGATTCCGCCATGTGCGAACCTCACGATTCTTCCACTGATAAGACCAGTGCTGATTATTACTTCGATTCATACT CTCACTTTGGTATTCATGAGGAAATGCTGAAGGATGTGGTGAGAACCAAGACTTATCAAAATGTCATCTACAAAAACAAA TTTTTATTCAAAGACAAGATTGTTCTTGACGTTGGTGCCGGGACCGGAATATTATCCCTCTTCTGTGCAAAAGCTGGGGC AAAACATGTTTATGCTATTGAGTGCTCACAGATGGCTGACATGGCACAAGAGATTGTTAAGGCGAACGGTTTTTCAAATG TTATAACTGTTTTGAAGGGAAAAGTTGAAGAAATCGACCTTCCAGTTCCACAAGTCGATATTATTATTTCGGAATGGATG GGTTACTTTCTGTTATACGAGAATATGTTGAATACAGTTTTATATGCTCGTGATAAGTGGCTGGTTAGCGATGGAATCGT GCTACCTGATAATGCAACACTTCATCTGACAGCAATAGAGGACTCCGAATACAAAGAGGACAAGATTGAATTTTGGAAAA ACGTGTACGGCTTTGACATGAGCTGCATCAGGAAACAGTCGATGATGGAACCTCTTGTTGACACAGTTGACCAGAATCAG ATCGTTACAAATTGTCAGTTACTTAAGAGCATGAATATCTCCAAGATGACGTCTGGAGATGCATCGTTTACAGCGCCTTT CAAGCTTGTGGCCGAGCGTGATGATTACATTCATGCTCTTGTAGCTTACTTTGATGTCGCTTTTACTGTTTGCCACAAAA TGACAGGATTCTCTACAGGGCCAAGGTCAAAGAATACACATTGGAAACAAACAGTGCTATATCTCGAAGATGTGTTAACT ATATGCCAAGGGGAGTTTGTGGTTGGAACCATGACAGTGACGCAAAACAAGAAAAATCCCCGAGACATTGACATCACACT CAAATACTCATTAAACGGTCGTCGTTGCAATGTGTCGAGGACCCAAGAATACAAGATGCGTTGA |