
| Microexon ID | Gm_7:15330531-15330541:+ |
| Species | Glycine max | Coordinates | 7:15330531..15330541 |
| Microexon Cluster ID | MEP26 |
| Size | 11 |
| Phase | 1 |
| Pfam Domain Motif | PRMT5 |
| Structure of Microexon-tag (flanking exon, microexon, flanking exon sizes) | 49,11,48 |
| Microexon location in the Microexon-tag | 2 |
| Microexon-tag DNA Seq | RRYGAYAARACCAGYGCYGAYTAYTACTTCGAYTCCTACTCYCAYTTYGGTATTCATGAAGAAATGYTGAARGATRYWGTGAGRACWAARACWTAYCAAAATGTTATY |
| Logo of Microexon-tag DNA Seq | 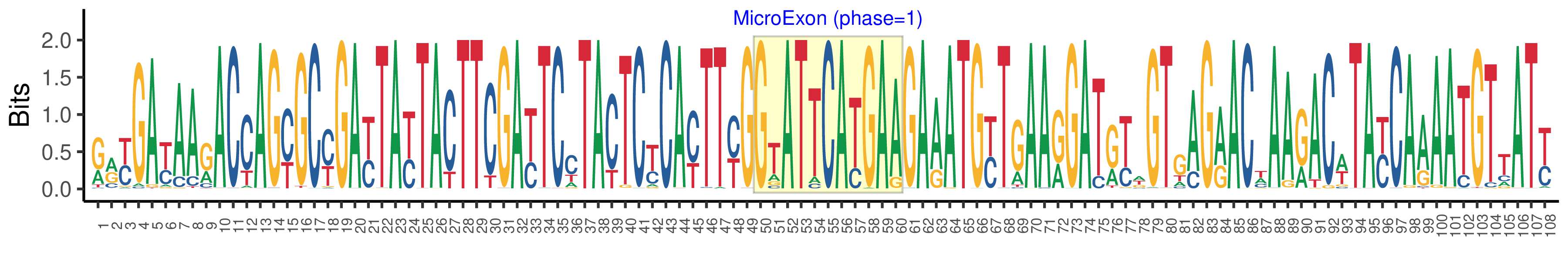 |
| Alignment of exons | 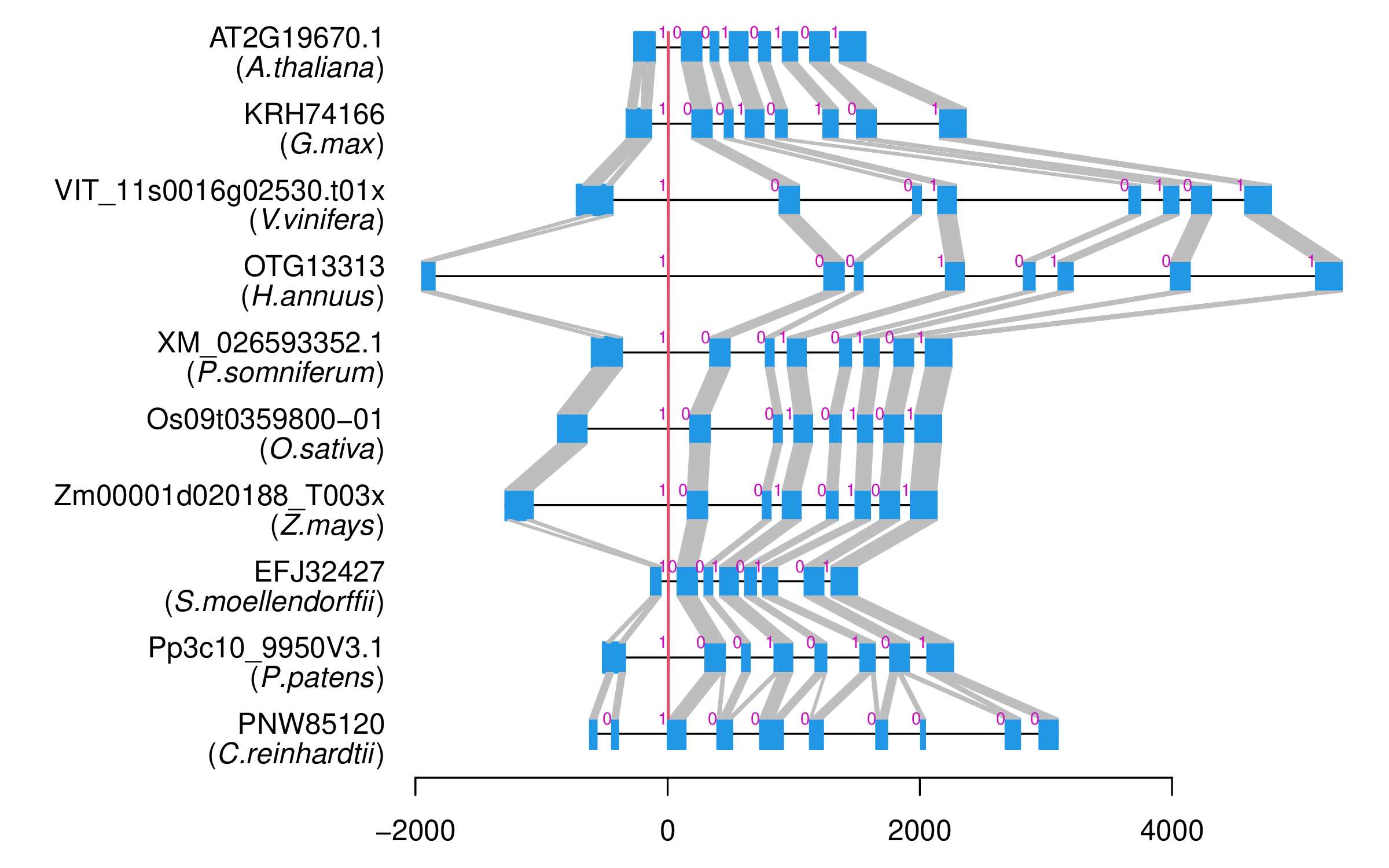 |
| Microexon DNA seq | GAATTCACGAA |
| Microexon Amino Acid seq | GIHE |
| Microexon-tag DNA Seq | GATGATAAAACCAGCGCCGACTATTACTTCGATTCCTACTCTCATTTTGGAATTCACGAAGAAATGTTGAAGGACAGTGTGAGAACCAAGACATATCAAAATGTTATT |
| Microexon-tag Amino Acid Seq | DDKTSADYYFDSYSHFGIHEEMLKDSVRTKTYQNVI |
| Microexon-tag spanning region | 15330359-15330765 |
| Microexon-tag prediction score | 0.9787 |
| Overlapped with the annotated transcript (%) | 100 |
| New Transcript ID | KRH49040x |
| Reference Transcript ID | KRH49040 |
| Gene ID | GLYMA_07G128100 |
| Gene Name | NA |
| Transcript ID | KRH49040 |
| Protein ID | KRH49040 |
| Gene ID | GLYMA_07G128100 |
| Gene Name | NA |
| Pfam domain motif | PRMT5 |
| Motif E-value | 0.00015 |
| Motif start | 73 |
| Motif end | 175 |
| Protein seq | >KRH49040 MGRRKNNSNNNQCSSSKEDADMNSNHLRFEEADEAVDESSNLDQSMCDIEESDDKTSADYYFDSYSHFGIHEEMLKDSVR TKTYQNVIYQNKFLFKNKVVLDVGAGTGILSLFCAKAGAEHVYAVECSHMADMAKEIVEANGYSNVVTVLKGKIEEIELP VAKVDIIISEWMGYFLLFENMLNSVLYARDKWLVDGGVVLPDKASLHLTAIEDADYKEDKIEFWNNVYGFDMSCIKKQAI MEPLVDTVDQNQIATNCQLLKTMDISKMAPGDASFAAPFKLVAERDDYIHALVAYFDVSFTKCHKLMGFSTGPRSRATHW KQTVLYLEDVLTVCEGEAIVGSMTVAPNKKNPRDVDIMLKYSLNGRRCNVSRVQYYKMR* |
| CDS seq | >KRH49040 ATGGGTCGGCGAAAGAACAACAGCAACAACAATCAGTGCTCCTCAAGCAAGGAAGACGCTGACATGAATAGCAACCACCT TCGGTTTGAAGAAGCTGACGAGGCTGTCGACGAGAGTTCCAACCTCGACCAATCCATGTGCGACATCGAAGAATCCGATG ATAAAACCAGCGCCGACTATTACTTCGATTCCTACTCTCATTTTGGAATTCACGAAGAAATGTTGAAGGACAGTGTGAGA ACCAAGACATATCAAAATGTTATTTATCAGAACAAATTTTTATTCAAGAATAAAGTAGTTCTTGATGTTGGTGCTGGGAC TGGGATTTTATCACTATTTTGTGCCAAAGCAGGGGCAGAACACGTCTATGCGGTTGAGTGCTCCCATATGGCTGACATGG CGAAGGAGATTGTTGAAGCTAACGGTTACTCTAATGTTGTAACAGTTTTGAAGGGGAAGATTGAAGAAATTGAACTTCCA GTTGCTAAAGTTGATATAATTATTTCAGAATGGATGGGATATTTCTTGTTGTTTGAGAATATGTTAAATTCTGTGCTCTA TGCTCGTGACAAATGGCTTGTGGATGGTGGAGTTGTGCTACCAGACAAAGCATCCCTACATCTCACTGCTATTGAAGATG CTGACTATAAAGAAGATAAAATTGAGTTTTGGAACAATGTATATGGATTTGACATGAGCTGCATCAAGAAGCAAGCCATA ATGGAGCCTCTTGTTGACACAGTTGACCAGAATCAGATTGCTACAAACTGCCAGCTACTCAAGACAATGGATATCTCAAA GATGGCTCCTGGAGATGCTTCATTCGCAGCACCTTTTAAGCTTGTAGCTGAACGCGACGACTATATTCACGCTCTTGTTG CATATTTTGATGTATCATTTACAAAGTGTCATAAATTGATGGGATTCTCTACAGGGCCAAGATCACGAGCTACACATTGG AAACAAACAGTCCTATACCTAGAAGATGTCTTGACCGTTTGTGAGGGGGAGGCAATTGTGGGGAGCATGACTGTTGCTCC AAATAAAAAAAATCCTCGGGATGTTGATATAATGCTCAAGTATTCATTGAATGGAAGGCGATGCAATGTTTCAAGGGTTC AGTACTACAAGATGCGTTGA |
| Microexon DNA seq | GAATTCACGAA |
| Microexon Amino Acid seq | GIHE |
| Microexon-tag DNA Seq | GATGATAAAACCAGCGCCGACTATTACTTCGATTCCTACTCTCATTTTGGAATTCACGAAGAAATGTTGAAGGACAGTGTGAGAACCAAGACATATCAAAATGTTATT |
| Microexon-tag Amino Acid seq | DDKTSADYYFDSYSHFGIHEEMLKDSVRTKTYQNVI |
| Transcript ID | KRH49040 |
| Gene ID | Gm.46762 |
| Gene Name | NA |
| Pfam domain motif | PRMT5 |
| Motif E-value | 0.00015 |
| Motif start | 73 |
| Motif end | 175 |
| Protein seq | >KRH49040 MGRRKNNSNNNQCSSSKEDADMNSNHLRFEEADEAVDESSNLDQSMCDIEESDDKTSADYYFDSYSHFGIHEEMLKDSVR TKTYQNVIYQNKFLFKNKVVLDVGAGTGILSLFCAKAGAEHVYAVECSHMADMAKEIVEANGYSNVVTVLKGKIEEIELP VAKVDIIISEWMGYFLLFENMLNSVLYARDKWLVDGGVVLPDKASLHLTAIEDADYKEDKIEFWNNVYGFDMSCIKKQAI MEPLVDTVDQNQIATNCQLLKTMDISKMAPGDASFAAPFKLVAERDDYIHALVAYFDVSFTKCHKLMGFSTGPRSRATHW KQTVLYLEDVLTVCEGEAIVGSMTVAPNKKNPRDVDIMLKYSLNGRRCNVSRVQYYKMR* |
| CDS seq | >KRH49040 ATGGGTCGGCGAAAGAACAACAGCAACAACAATCAGTGCTCCTCAAGCAAGGAAGACGCTGACATGAATAGCAACCACCT TCGGTTTGAAGAAGCTGACGAGGCTGTCGACGAGAGTTCCAACCTCGACCAATCCATGTGCGACATCGAAGAATCCGATG ATAAAACCAGCGCCGACTATTACTTCGATTCCTACTCTCATTTTGGAATTCACGAAGAAATGTTGAAGGACAGTGTGAGA ACCAAGACATATCAAAATGTTATTTATCAGAACAAATTTTTATTCAAGAATAAAGTAGTTCTTGATGTTGGTGCTGGGAC TGGGATTTTATCACTATTTTGTGCCAAAGCAGGGGCAGAACACGTCTATGCGGTTGAGTGCTCCCATATGGCTGACATGG CGAAGGAGATTGTTGAAGCTAACGGTTACTCTAATGTTGTAACAGTTTTGAAGGGGAAGATTGAAGAAATTGAACTTCCA GTTGCTAAAGTTGATATAATTATTTCAGAATGGATGGGATATTTCTTGTTGTTTGAGAATATGTTAAATTCTGTGCTCTA TGCTCGTGACAAATGGCTTGTGGATGGTGGAGTTGTGCTACCAGACAAAGCATCCCTACATCTCACTGCTATTGAAGATG CTGACTATAAAGAAGATAAAATTGAGTTTTGGAACAATGTATATGGATTTGACATGAGCTGCATCAAGAAGCAAGCCATA ATGGAGCCTCTTGTTGACACAGTTGACCAGAATCAGATTGCTACAAACTGCCAGCTACTCAAGACAATGGATATCTCAAA GATGGCTCCTGGAGATGCTTCATTCGCAGCACCTTTTAAGCTTGTAGCTGAACGCGACGACTATATTCACGCTCTTGTTG CATATTTTGATGTATCATTTACAAAGTGTCATAAATTGATGGGATTCTCTACAGGGCCAAGATCACGAGCTACACATTGG AAACAAACAGTCCTATACCTAGAAGATGTCTTGACCGTTTGTGAGGGGGAGGCAATTGTGGGGAGCATGACTGTTGCTCC AAATAAAAAAAATCCTCGGGATGTTGATATAATGCTCAAGTATTCATTGAATGGAAGGCGATGCAATGTTTCAAGGGTTC AGTACTACAAGATGCGTTGA |